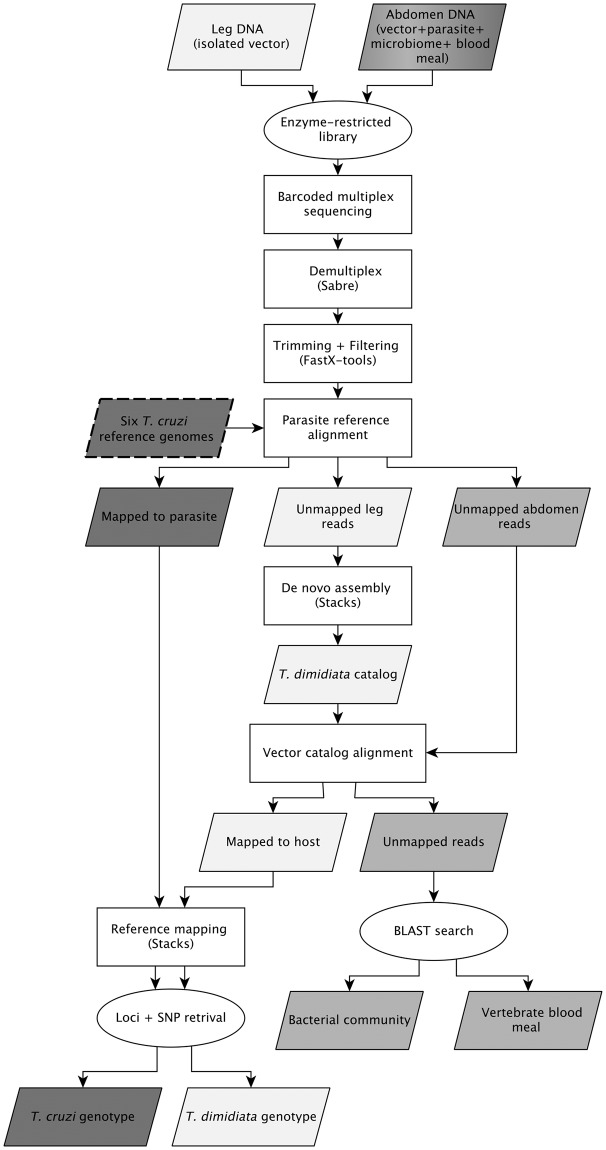
Fig 2. Bioinformatics pipeline separating RADseq data obtained from the legs and abdomens of Triatoma dimidiata specimens.
Raw data from 32 T. dimidiata were trimmed and filtered using FastX tools, then mapped to the six available T. cruzi genomes using Bowtie. The unmapped reads from the host were assembled denovo using Stacks, converted to an index, and used as a catalog to map all to T. dimidiata; both sets of mapped reads were aligned in STACKS to obtain markers for the parasite and host. The NCBI nt database was queried (May, 2016) with the remaining unmapped reads to quantify matches obtained from chordates (blood meal hosts), bacteria and other taxa. Input and output parallelograms are color-coded to indicate the vector (yellow), parasite (pink) and all other taxa (orange).