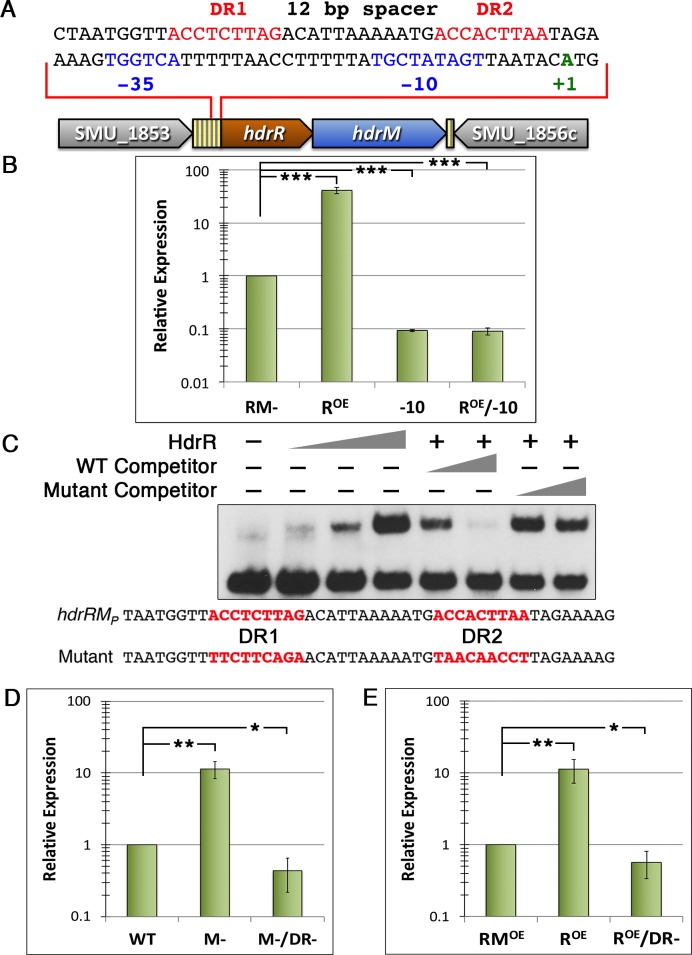
Fig 2. HdrRM operon regulatory elements mediate an autoregulatory positive feedback loop.
A) Partial sequence of the intergenic region upstream of the hdrRM ORFs. The operon promoter is shown in blue font, the direct repeats are shown in red font, and the hdrR operon transcription start site (+1) is shown in green font. ORFs are shaded in solid colors, whereas intergenic regions are striped. B) An hdrRM luciferase reporter was created by replacing the hdrRM ORFs with that of luciferase. The luciferase activity of this parent reporter strain (RM-) was then compared after ectopic hdrR overexpression (ROE), mutation of the operon promoter -10 site (-10), and after combining hdrR ectopic overexpression with a mutant operon promoter -10 site (ROE/-10). Data are presented relative to the parent reporter strain, which was arbitrarily assigned a value of 1. C) Electrophoretic mobility shift assays (EMSAs) were performed with recombinant HdrR and 1 ng of a labeled DNA probe (hdrRMP) encompassing the direct repeat region upstream of the hdrRM operon promoter. To confirm the specificity of HdrR binding to the direct repeats, an unlabeled wild-type DNA probe (hdrRMP) and an unlabeled direct repeat mutant DNA probe (Mutant) were added to the EMSA reactions as competitors. The sequences of both competitor probes are presented under the EMSA image with the direct repeats shown in red. HdrR abundance per reaction: Lane 1 (0 μg), Lane 2 (10 μg), Lane 3 (20 μg), and Lanes 4–8 (30 μg). Wild-type competitor DNA (hdrRMP) abundance per reaction: Lane 5 (50 ng) and Lane 6 (200 ng). Mutant competitor DNA (Mutant) abundance per reaction: Lane 7 (50 ng) and Lane 8 (200 ng). D) An hdrRM luciferase reporter was created by placing a luciferase ORF immediately downstream of the hdrRM ORFs. The luciferase activity of this parent reporter strain (WT) was then compared after mutating hdrM (M-) and after doubly mutating hdrM and the operon direct repeats (M-/DR-). Data are presented relative to the parent reporter strain, which was arbitrarily assigned a value of 1. E) An hdrRM luciferase reporter was created by replacing the hdrRM ORFs with that of luciferase and then ectopically overexpressing hdrR in a single copy on the chromosome, while hdrM was ectopically expressed from a multicopy plasmid (i.e. uncoupled hdrRM expression). The luciferase activity of this reporter strain (RMOE) was then compared to an hdrR ectopic overexpression reporter strain (ROE) and an hdrR ectopic overexpression reporter strain with mutated operon direct repeats (ROE/DR-). Data are presented relative to the reporter strain RMOE, which was arbitrarily assigned a value of 1. All luciferase data are expressed as means ± s.d. (indicated by error bars) derived from four biological replicates. ***P<0.001, **P<0.01, and *P<0.05, Unpaired two-tailed Student’s t-test with Welch’s correction, significance compared to RM- (B), WT (D), and RMOE (E).