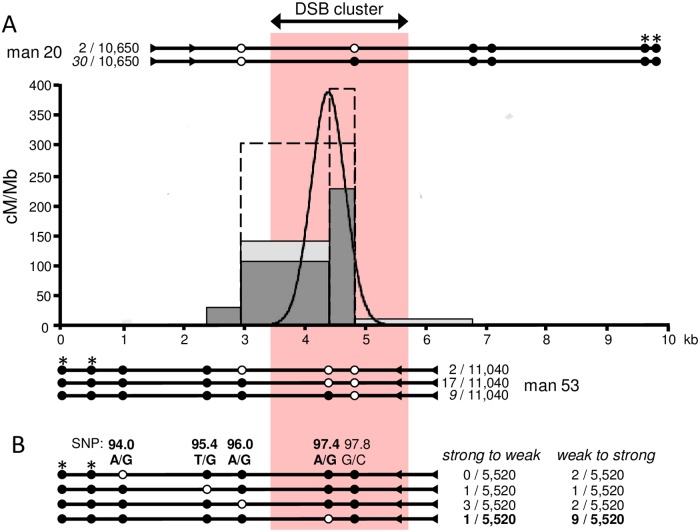
Fig 4. De novo recombination in the proximal region.
(A) Sperm CO profiles in relation to the proximal DSB cluster as determined by DMC1-SSDS [19] (pink panel) for man 20 (light grey histogram) and man 53 (dark grey histogram), with the combined least-squares best-fit normal distribution shown by the black curve. As for Fig 3, data from reciprocal assays have been pooled and the recovered structures and their frequencies for each man are shown above and below the histograms with informative SNPs represented by circles. In these assays, ASPs were designed against the SNPs marked with asterisks and were used in conjunction with universal primers (triangles) to selectively amplify each parental haplotype; recombinants were then detected by probing for the alleles of the opposite haplotype represented here by white circles (see Methods, S6 and S7 Tables). Note that CO events involving only the terminal marker closest to the universal primers are indistinguishable from NCO events in this assay, so we arbitrarily designated half of such events as COs in these cases (numbers given in italics) but indicate with dashed boxes in the graph how the profiles would appear should all such events actually be COs. In the latter case, the hotspot width would be reduced by 250 bp, the centre point would be shifted proximally by 116 bp, and the peak activity would be ~ 830 cM/Mb. (B) Testing for GC bias amongst NCOs. Of the four informative SNPs for man 53 that carry a ‘weak’ and ‘strong’ allele, SNP 97.4 shows over-transmission into NCOs of the ‘strong’ allele G relative to the ‘weak’ allele A (P = 0.011, one-tailed binomial exact test). This SNP lies 97 bp proximal to the hotspot centre as shown by the black curve in (A). Whilst we cannot be sure of the number of NCOs involving the terminal marker SNP 97.8, both alleles at this SNP base pair with three hydrogen bonds (i.e. are ‘strong’) and there is no evidence of disparity between the orientations assuming at least half the terminal recombinants are NCOs (i.e. 9). For man 20, terminal marker SNP 96.0 recombinants were recovered in the two orientations with similar frequencies, again suggesting an absence of TD. Note a further two NCOs each affecting a different single site (SNP 99.8 or SNP 100.1) were also recovered for this man but are not depicted in this figure.