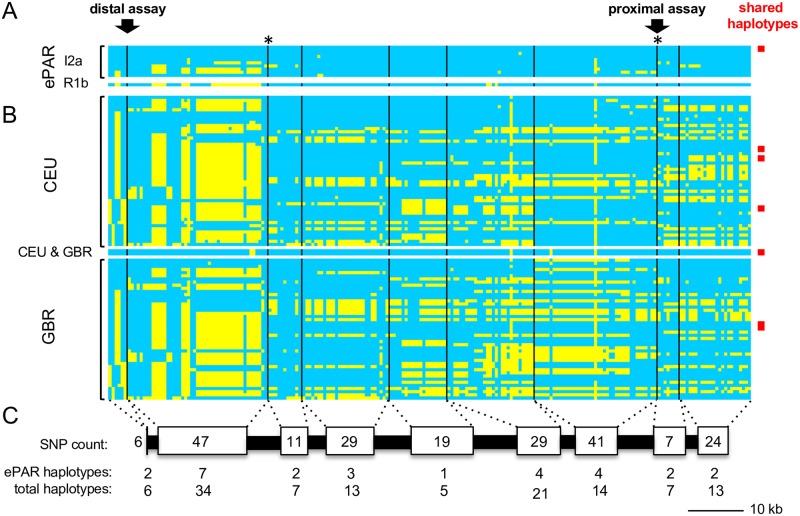
Fig 5. Comparison of inferred ePAR haplotypes with phase-known haplotypes from the corresponding region of the X chromosome.
(A) SNP haplotypes from each of the eleven independently sampled ePARs (ten from the haplogroup I2a Y chromosome lineage and one from the R1b Y lineage) are shown in rows, clustered according to the distal haplotype block. Individual 6889_01 is shown at the top with all his alleles colour-coded blue; yellow denotes the alternative SNP alleles not carried by this man. Black vertical lines correspond to the relative locations of mapped PRDM9 A and C DSB clusters; in two instances, marked by asterisks, an A and C cluster lie in very close proximity. Arrows indicate the distal and proximal sperm recombination assay regions and the red box indicates a second ePAR that is identical to that of 6889_01. In total, nine of the ten ePAR haplotypes are unique to this dataset. (B) Phased X haplotypes from the 1000 Genomes Project [34] for the 49 CEU males and 46 GBR males. One haplotype is shared between the two sample sets as indicated. In addition, three pairs of identical X haplotypes were noted among the CEU and one X haplotype was found to be carried by three different GBR men (red boxes). In total, 42 of the 46 CEU haplotypes and 43 of the 44 GBR haplotypes are unique. None of these X haplotypes matches any of the ePAR haplotypes. (C) Relative scaling of the regions depicted together with summary count of the number of SNPs, number of ePAR haplotypes and the corresponding total number of haplotypes seen amongst the ePAR, CEU and GBR datasets per block of SNPs.