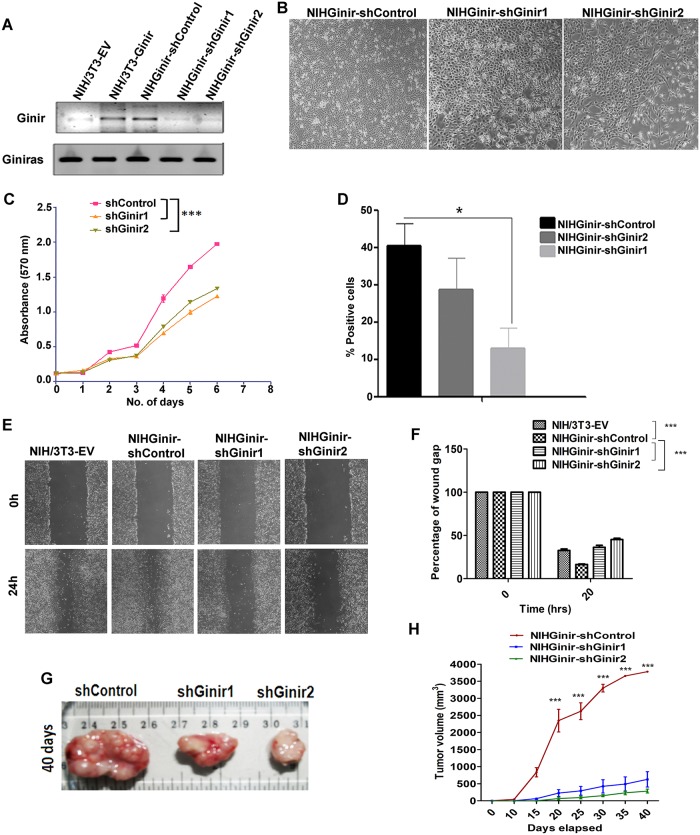
Fig 4. Down-regulation of Ginir RNA levels attenuates tumourigenic potential of NIH/3T3-Ginir cells.
(A) Representative RT-PCR analysis demonstrating Ginir levels in NIH/3T3-EV and NIH/3T3-Ginir(A) cells expressing Ginir-shRNA 1 and 2 and shControl (Scrambled) using G2F-G2R primers. Gapdh served as an internal RNA control. (B) Representative-phase contrast images of NIHGinir cells stably transfected with shRNAs to Ginir (shGinir1 or shGinir2) or with a scrambled shRNA (shControl). (Magnification 10×). (C) Cell proliferation analysis using MTT assay for the indicated cell lines. Data are mean ± SEM. ***P ≤ 0.0001, one tailed, paired Student’s t test. (D) Quantification of Ki67 antigen expression shown as percentage of positively stained cells compared to total number of cells per field (number of fields counted were at least 10) in indicated cell lines. Values are mean ± SEM. **P ≤ 0.001. Two tailed, by one-way ANOVA test. (E and F). Wound healing assays performed with NIH/3T3, NIHGinir-shControl, and NIHGinir-shGinir (1 and 2) cells. The wound gaps were observed at two different time points: 0 hours and 24 hours post wound formation (n = 3) (E). Statistical analysis of wound closure in mentioned cell types wherein the percentage of wound gap measured at indicated time points is plotted and the values are represented as mean ± SEM; ***P < 0.0001 by regular two-way ANOVA test (F). (G) Representative photographs of tumours demonstrating the inhibitory effects of Ginir-shRNA on tumour growth potential of NIH/3T3-Ginir(A) cells. Xenograft growth was monitored in NOD/SCID mice at regular intervals of 5 days for up to 40 days. Later, mice were killed, and tumours were dissected out. The assay was performed at least 3 times (n = 3). (H) Tumour growth kinetics of the mentioned cell lines monitored over a period of 40 days. Data are mean ± SEM; ***P value ≤ 0.0001 by regular two-way ANOVA test. Supporting data for C, D, F, and H can be found in S2 Data. Gapdh, glyceride 3-phosphate dehydrogenase; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide; RT-PCR, reverse transcription polymerase chain reaction; shRNA, short hairpin RNA.