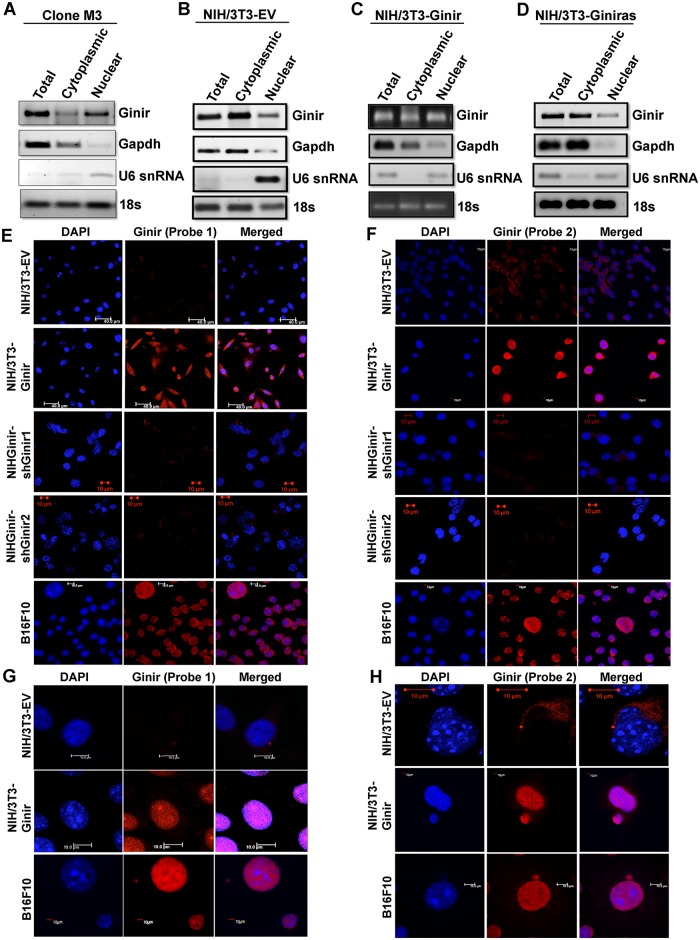
Fig 6. Ginir RNA is localised to cytoplasm in normal cells, but partitioning switches to the nuclear compartments in transformed cells.
(A-D) Subcellular fractionation followed by RT-PCR using G1F-G1R primers to determine compartmentalisation of Ginir and Giniras RNAs in (A) Clone M3, (B) NIH/3T3-EV, (C) NIH/3T3-Ginir, and (D) NIH/3T3-Giniras cells. Gapdh RNA served as an internal control for cytoplasmic RNA fractions and U6 snRNA as control for the nuclear fractions. The 18S rRNA served as a loading control for RT-PCR. A total of 35 cycles of amplifications were used for Ginir and Giniras RNAs, and 30 cycles were used for Gapdh RNA, U6 snRNA, and 18S rRNA. (E-H) RNA-FISH for Ginir localisation in the indicated cell lines using two independent LNA Ginir probes (Probe1-FAM labelled [E, G] and Probe2-Texas Red labelled [F, H]). Magnified images of a few cells are represented in G and H. Blue colour indicates nuclear staining with DAPI. Scale bar 10 μm. Gapdh, glyceride 3-phosphate dehydrogenase; Ginir, Genomic Instability Inducing RNA; Giniras, antisense RNA to Ginir; LNA, locked nucleic acid; RNA-FISH, RNA fluorescence in situ hybridisation; RT-PCR, reverse transcription polymerase chain reaction; snRNA, small nuclear RNA.