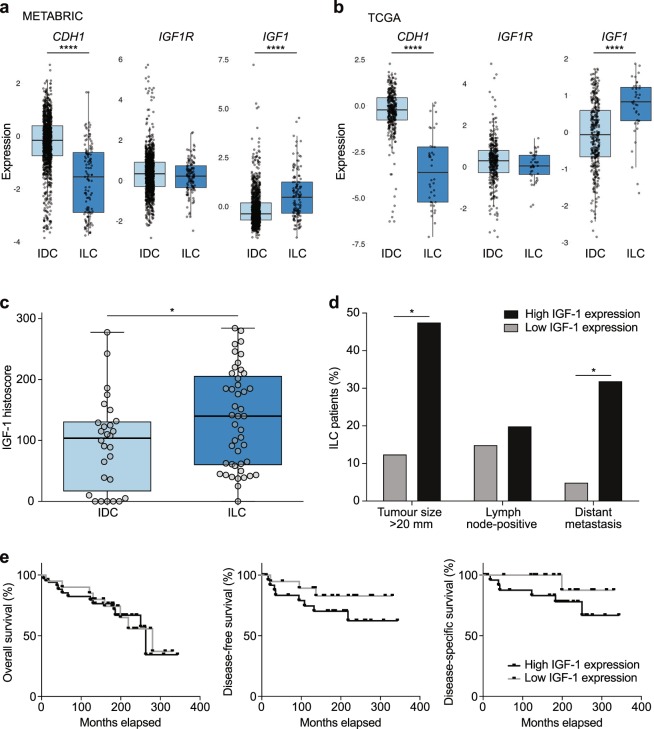
Figure 4.
IGF-1 expression is increased in human ILC versus IDC. (a,b) Boxplots of expression for CDH1, IGF1R and IGF1 genes from METABRIC (a) and TCGA (b) microarray mRNA expression datasets. All data points are ER-positive breast cancer samples. For further details, see Supplementary Table S5. Boxplots display the median (line), 25th and 75th percentiles (box) and 1.5 × interquartile range (whiskers). Light blue, IDC; dark blue, ILC. ****P < 0.0001; Wilcoxon test. (c,d) Analysis of IGF-1 cytoplasmic expression in a human TMA containing ILC and IDC samples. Boxplot (c) summarises IGF-1 histoscores. *P < 0.05; Mann–Whitney test. Histograms (d) show significant correlation between high cytoplasmic IGF-1 expression and increased tumour size and relapse with distant metastasis. *P < 0.05; tumour size, Pearson’s chi-squared test; distant metastasis, Fisher’s exact test. Lymph node status, P = 0.633; Fisher’s exact test. For further details, see Supplementary Table S6. (e) Kaplan–Meier plots representing proportions of patient survival. Disease-free survival was defined as time from primary surgery to first occurrence of relapsed disease (loco-regional recurrence and/or distant metastasis), and disease-specific survival was defined as time from primary surgery to breast cancer-related death. Overall survival (left panel), P = 0.869; disease-free survival (middle panel), P = 0.210; disease-specific survival (right panel), P = 0.135; log-rank test.