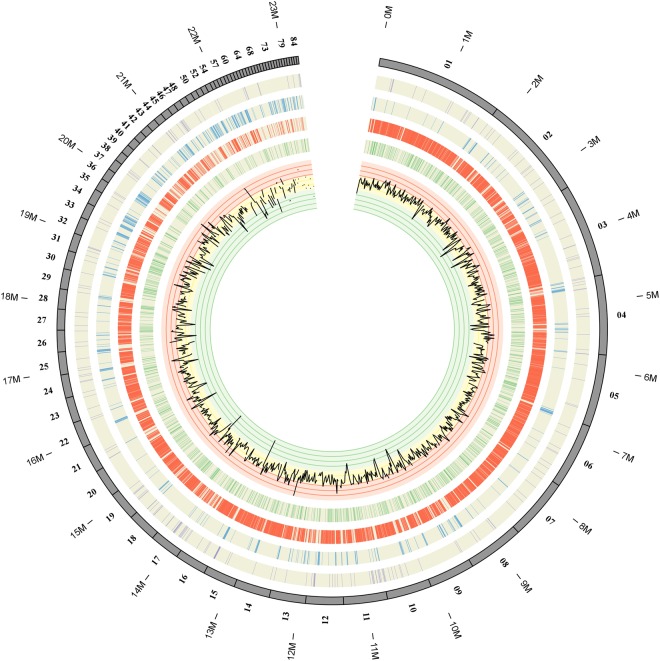
Figure 7.
Physical locations of predicted secreted protein genes, all protein-coding genes and PHI-base genes in relation to regions of repetitive sequences and GC content distribution in the assembled genome of T. horrida. From outside to in: Location of genes encoding predicted secreted proteins (purple) in the assembled genome; The distribution of transposable elements (blue) in the T. horrida genome; Single-copy DNA regions (red) of the T. horrida genome; Locations of the PHI-base gene homologues (green) involved in pathogen–host interactions; Graphs of GC (black) contents. Areas of low GC correspond well to regions of repetitive DNA. The maps were drawn with OmniMapFree.