Abstract
Respiratory complex I is part of a large family of homologous enzymes that carry out the transfer of electrons between soluble cytoplasmic electron carriers and membrane-bound electron carriers. These complexes are vital bioenergetic enzymes that serve as the entry points into electron transport chains for a wide variety of microbial metabolisms, and electron transfer is coupled to proton translocation. The core complex of this enzyme is made up of 11 protein subunits, with three major proton pumping subunits. Here, we document a large number of modified complex I gene cassettes found in genome sequences from diverse cultured bacteria, shotgun metagenomics, and environmentally derived archaeal fosmids all of which encode a fourth proton pumping subunit. The incorporation of this extra subunit into a functional protein complex is supported by large amino acid insertions in the amphipathic helix that runs the length of the protein complex. Phylogenetic analyses reveal that these modified complexes appear to have arisen independently multiple times in a remarkable case of convergent molecular evolution. From an energetic perspective, we hypothesize that this modification on the canonical complex I architecture allows for the translocation of a fifth proton per reaction cycle—the physiological utility of this modified complex is discussed.
Introduction
The generation and maintenance of an electrochemical proton gradient across the cytoplasmic membrane is centrally important for nearly all cells [1]. The energy stored in this gradient is used for ATP generation as well as the active transport of a range of metabolites and other ions to and from the cytoplasm through the action of a wide array of symporters and antiporters. Membrane-bound electron transport in respiring organisms is carried out in order to produce and maintain this gradient. Complex I, so named due to its position in the mitochondrial electron transport chain, is one of the most broadly distributed electron transport complexes in biology. Complex I homologs are capable of interacting with widely utilized cytoplasmic electron carriers such as NADH, F420H2 [2], flavodoxins [3], and likely ferredoxins [4, 5], depositing their electrons onto many diverse membrane-soluble electron carriers, including ubiquinone, menaquinone, and methanophenazine. Members of this family are often named based on the identity of the electron donors and acceptors such as NADH:ubiquinone oxidoreductase (Nuo), NADH:quinone oxidoreductase (Nqo), NADH dehydrogenase (Ndh), F420:methanophenazine oxidoreductase (Fpo), and F420H2:quinone oxidoreductase (Fqo). This substrate versatility is due to the modular nature of the complex, where the electron input and output modules can be adapted or replaced to facilitate new interactions, while keeping the core function of redox powered proton pumping intact.
Understanding of complex I function has benefited greatly from recent high-resolution crystal structures from thermophilic bacteria and yeast mitochondria [6–8] as well as single-particle electron cryo-microscopy of mammalian mitochondria [9]. These studies reveal significant similarities shared between distantly related complexes (reviewed in ref. [10]). Complex I consists of three modules that are summarized briefly here. The Nuo naming scheme is used throughout which assigns the names NuoA-N to genes in the order they appear in the operon of the 14 subunit complex that oxidizes NADH and reduces ubiquinone. NuoEFG form the cytoplasmic N-module that interacts with NADH and facilitates its oxidation (Fig. 1a). The NuoF protein binds an FMN cofactor, which serves as an intermediary between the 2-electron carrying NADH and 1-electron carrying iron–sulfur clusters that transfer the electrons through the complex. The NuoG protein contains multiple iron–sulfur clusters, and is homologous to members of the complex iron–sulfur molybdopterin (CISM) superfamily of proteins, although it does not bind the bis(MGD)Mo cofactors generally required for catalytic activity in the CISM superfamily [11].
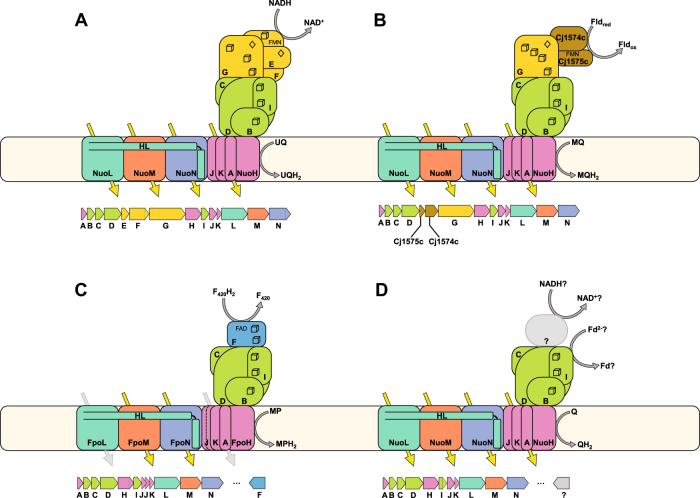
Fig. 1.
Common complex I homolog quaternary structure and gene order. Yellow arrows indicate proton translocations from the cytoplasm to the periplasm through the membrane-bound P-module. The C-terminal amphipathic helix of NuoL is labeled HL. 4Fe4S sulfur clusters are depicted as cubes, 2Fe2S clusters as diamonds. a Canonical 14 subunit NADH:ubiquinone oxidoreductase (Nuo) as found in T. thermophilus. Genes are generally arranged in an operon in alphabetical order as depicted. b A complex I cassette found in Camplylobacterota and best characterized in C. jejuni and H. pylori. The NuoEF genes are replaced by two smaller, non-homologous, proteins, which facilitate the interaction with flavodoxin (Fld) instead of NADH. The NuoG proteins in these operons contain an extra iron–sulfur cluster. c A methanogen-type F420H2:methanophenazine oxidoreductase (Fpo) showing the replacement of the N-module (NuoEFG) with the FpoF gene (in blue). This gene is not commonly associated with the rest of the operon, but its inclusion in the functional protein complex has been demonstrated by purification of the native Fpo complex. Note: two genes are annotated as NuoJ in these operons, but this represents a fission of the canonical NuoJ gene, and not a gene duplication event. d A common complex I gene cassette found in many organisms containing only the core 11 subunits of the complex. Few of these complexes have been studied biochemically and it is currently unknown whether the electron donors for these complexes are soluble proteinaceous electron carriers such as ferredoxins (Fd), or whether additional proteins encoded elsewhere in the genome are recruited to the complex to interact with small molecule electron carriers like NADH
In many Campylobacterota [12] such as Campylobacter jejuni and Helicobacter pylori the complex I operons contain NuoG but lack the NuoEF genes and have instead incorporated two unrelated protein subunits in their place (Fig. 1b). These two subunits allow these complex I homologs to use a flavodoxin protein as the electron donor instead of NADH [3]. The N-module is absent in archaea where it is sometimes functionally replaced by FpoF—a subunit that binds an FAD cofactor and oxidizes a lower potential electron carrier, F420H2 [2, 13] (Fig. 1c). FpoF is not often associated with the rest of the Fpo gene cluster. Many other bacterial and archaeal complex I operons also lack the N-module, with no replacement contained in the gene cluster, leaving the identity of the physiological electron donor in question [14] (Fig. 1d). Hypotheses for the electron donor for these complexes include the recruitment of other proteins to functionally replace the diaphorase activity of the N-module, as was hypothesized for the Hox hydrogenase [15] and NdhV [16] in Cyanobacteria, FAD/NAD(P) binding oxidoreductases in ammonia oxidizing Thaumarchaea [17], or the complete absence of an N-module analog, and instead the direct interaction between the complex I homolog and soluble proteinaceous electron carriers such as ferredoxins [18].
The Q-module is comprised of subunits NuoBCDI that together bind additional iron–sulfur clusters and accept electrons from the N-module; they also partially facilitate the reduction of a quinone bound on the cytoplasmic face of the membrane. NuoD is homologous to the large catalytic subunits of NiFe hydrogenases but, in an interesting evolutionary parallel to NuoG, appear to have lost the residues required to bind the active site cofactors.
The remaining subunits NuoAHJKLMN are all integral membrane proteins referred to as the P-module. NuoL, M, and N pump one proton each during a reaction cycle, with a final fourth proton being pumped by the combined action of the remaining NuoAHJK subunits [10, 19]. NuoL, M, and N are homologous to one another as well as to subunits from sodium-proton antiporters, formate-hydrogen lyases, carbon monoxide dehydrogenases, and membrane bound hydrogenases—relationships that reveal the shared evolutionary history of many major types of bioenergetic proton pumping complexes [20]. NuoL contains a unique structural feature that sets it apart from NuoM and NuoN: a C-terminal extension that forms an amphipathic helix reaching from the far end of the complex back to the NuoN subunit [10] (illustrated in Fig. 1). It is thought that this helix is important for transducing the energy released in the redox reactions of the Q-module into conformational changes in the P-module, leading to proton translocation. This helix has been described as a molecular “piston” [6], and site-directed mutagenesis studies creating insertions or deletions in this helix have deleterious effects on complex I assembly and function [21, 22]. The Fpo complex of methanogenic archaea may pump fewer protons based on biochemical measurements of the H+/e− stoichiometry, which are consistent with the less favorable energetics of the F420/methanophenazine redox couple [23] (Fig. 1c) and sequence analysis of the FpoL gene from a variety of methanogenic archaea [24], although this finding has yet to be confirmed with purified protein complexes.
Here we describe multiple occurrences and evolutionary histories of gene cassettes encoding complex I homologs that incorporate a second full-length copy of the NuoM subunit. These modified complex I homologs can be found in many phylogenetically diverse organisms, and appear to have arisen through convergent evolution involving the horizontal gene transfer of a second, distantly related copy of NuoM. Importantly, comparisons of the primary amino acid sequences of the peptides encoded by these gene cassettes supports the proposal that this extra NuoM is incorporated into a single-functional complex by modifications that lengthen the NuoL amphipathic helix. The inclusion of the additional subunit suggests the upgrading of these complexes to a 5 H+/2e− stoichiometry, which if correct would reflect the most protons pumped per reaction cycle by any bioenergetic complex.
Materials and methods
Complex I subunit gene selection
A rarified subset of microbial genomes (1860) were selected from nearly 26,000 archaeal and bacterial genomes available on the IMG [25] server in April 2015. The rarified list was selected manually, keeping approximately one genome from each prokaryotic genus. Finalized genomes were favored over partial or draft genomes when available. Initial complex I gene homologs were selected by retrieving all genes from this genome set automatically annotated as one of the Nuo genes by Kegg Ortholog number (see below). In our experience these proteins tend to be well annotated, although there are occasional contaminating proteins from other bioenergetics subunits, which were manually excluded from the analysis if they did not fall in what appeared to be a complex I operon. After identifying complex I clusters with additional pumping subunits, additional sequences from close relatives of those complexes were retrieved from databases including NCBI and IMG/M.
Protein sequence alignment
All proteins annotated as NuoL (including NuoL, NuoLM, and NdhF [K00341, K15863, and K05577]) and NuoM (including NuoM, NuoLM, and NdhD [K00342, K15863, and K05575]) were separately aligned with Clustal Omega [26], MUSCLE [27] and MAFFT [28]. Many of the loops between transmembrane helices in NuoL and NuoM have lineage-specific indels that are not homologous features, and confound phylogenetic reconstruction, so we utilized GBlocks to identify well-conserved positions in our alignments that were suitable for building phylogenies [29]. GBlocks was run on the Castresana Lab web server with low stringency settings allowing for gap positions, less strict flanking positions and smaller final blocks. The resulting alignments consisted of 274, 249, and 232 positions for NuoL from Clustal Omega, MAFFT, and MUSCLE, respectively; 201, 169, and 118 positions for NuoM, from Clustal Omega, MAFFT, and MUSCLE, respectively; and 201, 154, and 145 positions for NuoN, from Clustal Omega, MAFFT, and MUSCLE, respectively. We rarified the dataset using a 90% identity cutoff to reduce the dataset to a manageable size for phylogenetic reconstruction. After rarification the datasets contained between ~650 and ~1100 sequences depending on the subunit and alignment method used. Datasets were further rarified using 80% and 70% identity cutoff values for testing different phylogenetic models.
Phylogenetic reconstruction
Trees were constructed with RAxML and MrBayes. RAxML trees were built with RAxML-HPC v.8 on the CIPRES Science Gateway V 3.3 [30] with 25 distinct rate categories, CAT or GAMMA models, and with LG, WAG or Dayhoff substitution matrices for each of the three alignments described above. Only the Clustal aligned trees using the 90% identity dataset with CAT and LG are shown in main text figures, those based on the other two alignment methods and setting variations are included as Supplementary figures. MrBayes trees were built with MrBayes v3.2.6 on CIPRES for only the Clustal alignment and LG substitution matrix due to the computational demand of the MrBayes program. Clade-specific trees were constructed with RAxML LG + CAT on all three aforementioned alignment programs without GBlocks.
Structural homology modeling
NuoL genes from each clade and the NuoG gene from Nitrospira defluvii were submitted to the I-TASSER web server [31–33]. For Clade I the NuoL sequence from Chloroflexus sp. Y-400-fl was submitted and the top model had a C-score of 0.4, TM-score 0.77 ± 0.1 and RMSD of 7.3 ± 4.2. For Clade 2 the NuoL sequence from Desulfonatronovibrio magnus was submitted and the top model had a C-score of 0.82, TM-score 0.82 ± 0.08 and RMSD of 6.1 ± 3.8. For Clade 3 the NuoL sequence from Nitrospira defluvii was submitted and the top model had a C-score of 0.62, TM-score 0.8 ± 0.09 and RMSD of 6.7 ± 4. The top model for N. defluvii NuoG had a C-score of 0.08, TM-score 0.72 ± 0.11 and RMSD of 8.5 ± 4.5.
Genome sequencing and assembly
The genomes of Bellilinea caldifistulae GOMI-1 (DSM 17877) and Literilinea aerophila PRI-4131 (DSM 25763) were sequenced as part of a project to expand the phylogenetic breadth of Chloroflexi genomes. Genomic DNA was ordered from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) and sequenced using the Illumina MiSeq platform. SPAdes 3.1.1 [34] was used to assemble the genomes. For B. caldifistulae, sequence coverage, GC composition and phylogenetic affiliation of conserved single copy genes were used to exclude contaminating contigs and genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline. The draft genome is 3.72 Mb in size, comprised of 31 contigs, 3399 genes, 2990 CDSs, and is estimated to be ~95% complete based on conserved single copy genes (106/111). This whole-genome shotgun project has been deposited in DDBJ/EMBL/GenBank under the accession number LGHJ00000000. L. aerophila sequencing data were not high enough quality to produce a draft genome, however, SPAdes assembly did produce a contig containing the complex I gene cassette. The DNA sequence for this contig is deposited in NCBI under accession number JWSX01002273.1.
Results and discussion
Diverse complex I gene cassettes with extra proton pumping subunits
Through detailed investigation of the diversity of complex I homologs we discovered a unique type of complex I gene cassette encoding a second full-length copy of the large proton pumping subunit NuoM. These gene clusters (herein referred to as 2M complexes) are observed in genomic data from five different bacterial phyla, environmentally derived Thaumarchaeal fosmids, and a number of genome bins from assembled metagenomes. Examples of 2M complex gene clusters are shown in Fig. 2a, and were initially classified into three clades according to their gene synteny and primary sequence similarity. These gene cassettes never included the NuoEF genes, and in all cases were found in genomes that contained canonical complex I operons elsewhere in the genome.
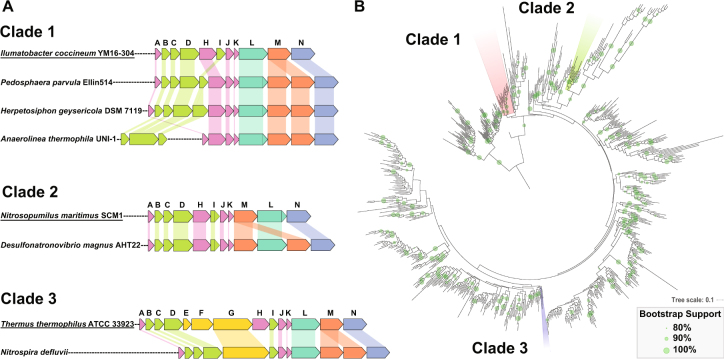
Fig. 2.
2M complex I homologs were found in three discrete clades. a Examples of the operon structure of 2M complexes from each of the three different clades showing the addition of a second NuoM subunit (in orange), as well as general operon features. Closely related canonical complex I operons are shown for comparison (underlined). In Clade 1 the second NuoM occurs right after the first, and the operons contain a unique gene rearrangement that swaps the NuoH and NuoI subunits; in some members of this Clade such as Anaerolinea thermophila UNI-1 the NuoBCDI genes occur separately from the rest of the operon. In Clade 2 the two NuoM genes are found on either side of NuoL; the NuoM preceding the NuoL is characteristic of the complex I operons found in Thaumarchaea. None of the operons have full N-modules, and only Clade 3 has a NuoG homolog in its operon. b A maximum likelihood phylogenetic tree of NuoL subunits from a rarified subset of publically available genome data. The NuoL genes from Clades 1, 2, and 3 fall far from one another in well-supported groups surrounded by NuoL genes in normal complex I operons—a pattern that implies that multiple independent events of convergent evolution gave rise to the 2M complex Is. Bootstrap support is shown for values ≥80%. High resolution NuoL trees, including those built from MUSCLE and MAFFT alignments, are shown in Figures S1-10
Phylogenetic trees built from NuoL homologs from a broad sampling of publically available data reveal that the three clades of 2M complexes form phylogenetically coherent groups comprised of organisms from: Clade (1) Chloroflexi, Verrucomicrobia and Acidobacteria; Clade (2) uncultured Thaumarchaea and Deltaproteobacteria of the Desulfovibrionales order; and Clade (3) the genus Nitrospira (Fig. 2b). In each case the most closely related NuoL proteins compared to those found in the 2M clades occurred in canonical complex I operons without the second NuoM subunit (as in Fig. 2a). This suggests three separate evolutionary events in which a second NuoM was added to a canonical complex I operon. The accuracy of these phylogenetic relationships were evaluated using three primary sequence alignment tools, three different substitution matrices and two different rate heterogeneity models—all yielding phylogenies confirming the polyphyletic nature of the 2M clades (Methods section, Figures S1-10).
To determine the origins of these second NuoM subunits contained in the 2M operons we constructed phylogenetic trees from a broad sampling of NuoM subunits in a similar manner as NuoL. As was observed with the NuoL phylogeny, all three clades were distinct from one another in the NuoM tree (Fig. 3a). However, the two copies of NuoM from each clade are not closely related to one another. This illustrates that the second NuoM was not derived from direct duplication of NuoM from within the operon, but instead was incorporated from a separate complex I homolog through horizontal gene transfer. Interestingly the NuoM2 gene from the Clade 2 Desulfovibrionales is located within a completely different region of the tree than the NuoM2 from the Thaumarchaeal fosmids of this clade. It appears that although the NuoL subunits in Clade 2 are all closely related, the additional NuoM subunits were acquired independently in these two separate lineages. Phylogenies of the last large proton pumping subunit NuoN revealed the same polyphyletic pattern of the three 2M clades, including the separate clusters between the Desulfovibrionales and Thaumarchaea members of Clade 2 (Fig. 3b). Additional NuoM and NuoN trees were constructed using the same approach as with the NuoL trees, all yielded similar relationships (Figures S11-30). Interestingly, in some trees the NuoN genes from Clade 1 are split, with those from the Chloroflexi forming one group and the Acidobacteria and Verrucomicrobia forming a second. This was not true in all trees however, so the robustness of this observation requires further investigation. In no instance were the NuoL, NuoM, or NuoN genes in the canonical complex I operons from these organisms closely related to their homologs within the 2M complexes.
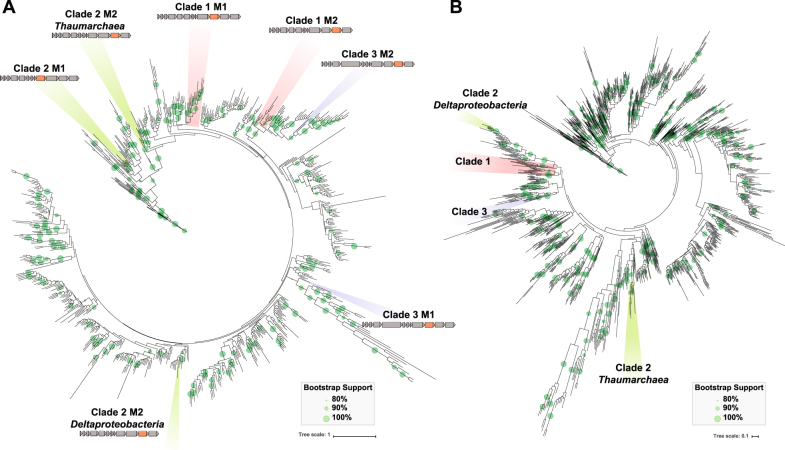
Fig. 3.
Phylogenetic trees of NuoM and N subunits from a rarified subset of publically available genome data. Bootstrap support is marked on the branches with values ≥80% support are shown. a As observed in the NuoL tree, the NuoM genes from Clades 1, 2, and 3 remain separate and distinct. Within Clade 2 the NuoM2 genes appear to be polyphyletic, with separate origins for the Thaumarchaeal NuoM2 and the Deltaproteobacterial NuoM2. Operon diagrams illustrate which NuoM homolog is found in each position of the tree. b The NuoN genes also show the polyphyletic nature of the three 2M clades, as well and separation between Deltaproteobacteria and Thaumarchaeal members of Clade 3. High resolution NuoM and N trees, including those built from MUSCLE and MAFFT alignments are shown in Figures S11-30
Evolution of 2M gene cassettes
To increase our understanding of the evolution and diversity of 2M complexes, we retrieved many additional members of the 2M clades to build clade-specific phylogenies. We also retrieved their nearest relatives from the NuoL tree in Fig. 2a to use as outgroups for our analysis. These additional 2M gene cassettes came from publically available data as well as our own sequencing efforts targeting diverse groups of cultured Chloroflexi (Methods section). The operon structures of this expanded collection of 2M complexes are shown in Fig. 4a–c and are ordinated by the phylogenetic relationships of their NuoL proteins. NuoL was chosen for reconstructing their phylogenetic history because it is the largest protein in the operon (~600 amino acids) and there is reason to believe that concatenating all of the genes together to make a composite phylogeny would be inappropriate due to the possibility that the individual genes in these operons have distinct evolutionary histories. In all three clades there is a high degree of congruence between the NuoL phylogeny and gene synteny in the operons suggesting the tree topology accurately captures some key aspects of the evolution of these gene clusters.
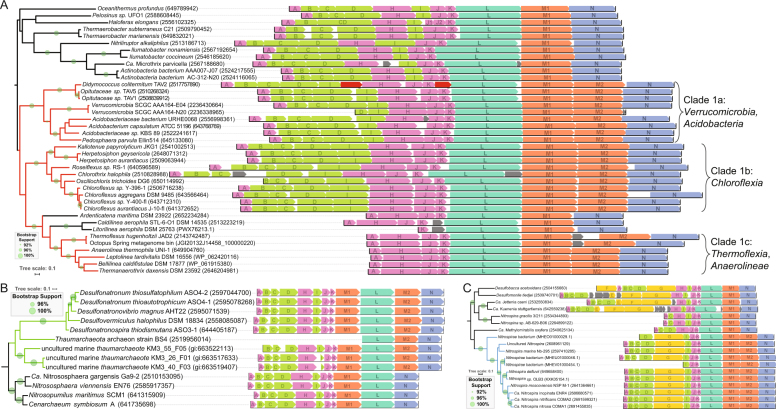
Fig. 4.
2M Clade-specific NuoL phylogenies with operon structures. a Clade 1 phylogeny reveals three major groups; 1a: Verrucomicrobia and Acidobacteria, 1b: Chloroflexia, 1c: Thermoflexia and Anaerolinea. All members and outgroups for Clade 1 lack NuoEFG genes and exhibit the unique reordering of the NuoH/I genes as highlighted in Fig. 2a. Possible mobile genetic elements in the Didymococcus colitermitum operon are highlighted in red. b Clade 2 is comprised of members of the Deltaproteobacteria and marine Thaumarchaea fosmids; it shares the NuoML gene order found in the other known Thaumarchaea used as an outgroup. c Clade 3 is comprised of members of the Nitrospira genus and contains a large NuoG gene as the only subunit of the N-module. The designations NuoM1 and NuoM2 were assigned based on which copy was more similar to the NuoM found in the most closely related normal complex I operons. Note: some operons are truncated due to their location on the ends of genomic contigs, and do not reflect segmentation or truncation in intact genomes
The 2M clades differ from one another in their operon structure and gene content. All members of Clades 1 and 3 have inserted the additional NuoM2 subunit between the original NuoM1 and NuoN subunits (Fig. 4a, c); whereas, the representatives from Clade 2 all have the gene order NuoMLMN, with the additional NuoM2 gene being inserted between the L and N (Fig. 4b). The gene order with a NuoM preceding NuoL is characteristic of Thaumarchaeal and Crenarchaeal complex I operons, which agrees with the phylogenetic placement of this group with the NuoL from canonical complex I operons in these archaea, as well as some members of Clade 2 being found in Thaumarchaeal fosmids [35].
In Clade 1 three large subgroups are apparent based on the phylogeny of NuoL, the phylogeny of the host organism, and the operon structure (Fig. 4a). Clade 1a contains examples found in Verrucomicrobia and Acidobacteria, Clade 1b contains members of the Chloroflexia class of the Chloroflexi and Clade 1c is found in Thermoflexia and Anaerolineae classes of the Chloroflexi. All members of Clade 1 lack the N-module genes and exhibit a unique gene order where all subunits of the Q-module (NuoBCDI) occur one after the other. This pattern never occurs in normal complex I operons, and might be a result of the gene rearrangement(s) that led to the incorporation of an additional NuoM gene. Notably, the ORFs marked in red in the Didymococcus colitermitum TAV2 operon are annotated as an IS200-like transposase and a member of the PD-(D/E)XK nuclease superfamily, which could be remnants of the mobile genetic elements responsible for the genome rearrangements that formed these 2M operons (Fig. 4a). In Clade 1c the NuoBCDI genes are found together in a separate part of the genome, with NuoC and NuoD fused (as depicted in Fig. 2a). This NuoCD fusion is not unique to 2M complexes and also exists in normal complex I operons [36], but the separation of the Q-module genes from the rest of the complex is peculiar.
Clade 3 operons from the Nitrospira genus have two important differences between them and the other 2M complexes. First, is the complete absence of a NuoH gene. As mentioned above, members of the Nitrospira have a canonical Nuo-type complex I however, so at least one copy of NuoH is present in these organisms; perhaps this NuoH is used by both complexes. No lone NuoH genes could be found elsewhere in any of the genomes. The second major difference is the presence of a large NuoG homolog in the gene cluster where the N-module proteins are normally located. As mentioned above, NuoG is part of the CISM superfamily, and in traditional complex I lacks the bis(MGD)Mo cofactor. However, the NuoG homolog in these gene cassettes is much more similar to members of the CISM superfamily than normal NuoG proteins, particularly the large catalytic subunits of formate dehydrogenases, so it is possible that in the Clade 3 2M complexes the NuoG has a novel catalytic role. This similarity to formate dehydrogenases extends to the incorporation of an additional iron–sulfur cluster, which connects the iron–sulfur clusters used by canonical NuoG to the ancestral CISM active site as has been previously reported for the NuoG proteins of H. pylori and C. jejuni [37]. However, one of the key ligand binding residues, SeCys/Cys140 (PDB:1FDO numbering) used for ligating the molybdenum atom in the bis(MGD)Mo cofactor [38], is replaced by a glycine in the Clade 3 NuoG homologs (Figure S31). The absence of this cysteine argues against the Clade 3 NuoG homologs being catalytically active in the known CISM fashion. The H. pylori and C. jejuni complex I gene cassettes lack NuoEF as well, but they have been replaced by two additional proteins that have resulted in the utilization of flavodoxin instead of NADH [3]. Neither of these two proteins have homologs in the Nitrospira genomes.
Extension of the NuoL amphipathic helix
The acquisition of a second copy of NuoM and its incorporation into a consistent well-ordered operon suggests that these 2M gene cassettes may produce larger versions of complex I with the ability to pump an extra proton per reaction cycle. To further investigate this idea, we closely examined the sequences of the NuoL genes from these 2M complexes in light of available crystal structure data. If these 2M complexes contain an additional NuoM subunit one would expect the entire P-module to be extended in length by the width of this subunit, and a canonical NuoL subunit positioned at the distal end of a 2M complex would not be able to reach its amphipathic helix past the second copy of NuoM all the way to the NuoN (see Fig. 1).
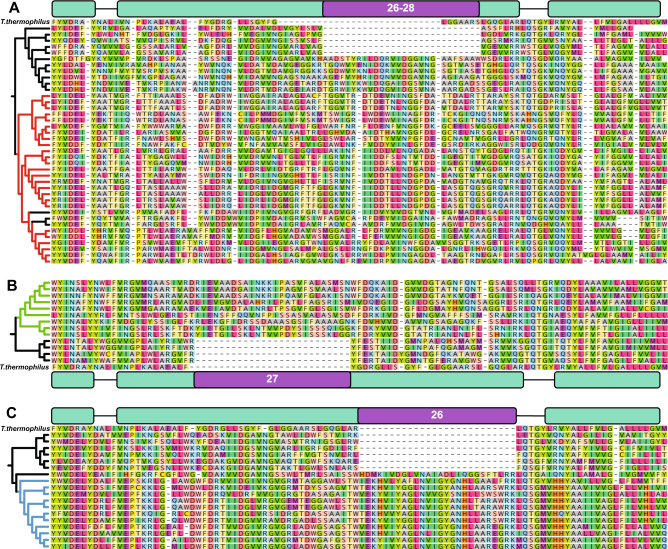
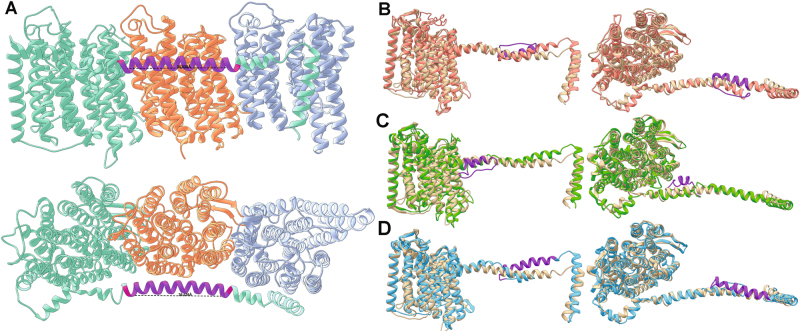
Remarkably, the alignments of the NuoL proteins in each of the three clades described above have an insertion of 26–28 amino acids in the middle of the amphipathic helix compared to their closest relatives lacking the second NuoM, as well as the NuoL sequence from T. thermophilus (Fig. 5a–c). Although estimates of the exact position of these insertions vary slightly based on the sequence alignment algorithm used, they all place an insertion of this size within the amphipathic helix (see Methods section, Figures S32-34). Using the T. thermophilus crystal structure, we measured the distance spanned by 26 amino acids along the amphipathic helix to be ca. 37 Å—a distance that nearly exactly matches the width of the NuoM subunit (Fig. 6a). Structural homology models built using I-TASSER [31–33] fit the NuoL subunits from all three 2M clades very well onto existing complex I crystal structures and have placed the extra 26–28 amino acids as insertions in the middle of the amphipathic helix in agreement with the multiple sequence alignments (Fig. 6b–d).
Fig. 5.
Sequence alignments highlighting the amphipathic helix insertions of the NuoL subunits from 2M complexes and their close relatives. Cladograms represent the same tree topology as those in Fig. 4, with colored branches marking the 2M groups. The T. thermophilus sequence is included in the alignment as a sequence length metric of where the amphipathic helix begins and ends. a Clade 1 contains 26–28 amino acids insertions compared to many of the close relatives and T. thermophilus. b Clade 2 contains 27 amino acid insertions compared to their closest relatives (28 compared to T. thermophilus). c Clade 3 contains 26 amino acid insertions. Notably, in a and c the closest relatives of the 2M complexes that were used as outgroups in the phylogenetic analysis also have insertions in the amphipathic helix, despite not having a second distantly related NuoM in their complex I operons. MUSCLE alignments are shown here, Clustal Omega and MAFFT alignments are shown in Figures S32-S34
Fig. 6.
Nuo crystal structure and structural homology models highlighting the characteristic amino acid insertion lengthening the amphipathic helical arm that spans the 2M complexes. a NuoL, M, and N subunits from the 4HEA crystal structure with a stretch of 26 amino acids marked in purple on the amphipathic helix (28 denoted by the additional red residues on either end of the purple region). This length of helix is approximately the same width as the NuoM subunit shown in orange. I-TASSER structural homology models of NuoL from Clades 1, 2, and 3 modeled onto the NuoL subunits from 4HEA are shown in b, c, and d, respectively. In all cases the insertions noted in Fig. 5 have been colored purple, and were modeled by I-TASSER as helices that doubled back on the original helix
Some of the closest relatives of the 2M complex NuoL genes contain the amphipathic helix insertion, but lack the additional NuoM. In Clade 1, Caldilinea aerophilum, Ardenticatena maritima, Litorilinea aerophila, and five members of the Actinobacteria outgroup complex all have the insertion (Fig. 5a). No close relatives of Clade 2 exhibited this insertion, but the closest relative of the Clade 3 complexes, Ca. Methylomirabilis oxyfera, had an insertion without a second NuoM as well (Fig. 5c). From this we interpret the insertion of the 26–28 amino acids in the NuoL amphipathic helix as a potentiating mutation that enabled the addition of a second, distantly related NuoM subunit. It is worth noting that many of the Actinobacteria (Ca. Microthrix parvicella, Nitriliruptor alkaliphilus, AAA007-J07 and AC-312-N20), C. aerophila, and Ca. M. oxyfera all have a second complex I gene cassette immediately up or downstream of the one depicted in Fig. 5, raising the possibility that these organisms might use an additional NuoM from the second complex in lieu of forming their own 2M operon. However, the NuoM from these complex I gene cassettes are not closely related to the NuoM2 found in the 2M complexes. It also remains a possibility that in these operons containing an extended NuoL two copies of the same NuoM protein are used to form a 2M protein complex.
Bioenergetic considerations and physiological conditions leading to increased proton pumping
The physiological and ecological utility of these novel 2M complexes remains to be discovered, however multiple instances of their convergent evolution, combined with their diversity and cross-domain horizontal gene transfer suggests that they have meaningful adaptive value. We speculated that by containing an extra proton-pumping subunit, 2M complexes achieve a higher stoichiometry of protons translocated per 2e− reaction cycle. On its face, such a trait might seem like an advantage for all metabolisms that employ complex I, but to inform this idea it is useful to examine the energetics of the reactions facilitated by these enzymes. From a thermodynamic point of view, complex I and its homologs convert energy released from a redox reaction into transmembrane proton motive force. Below we examine how the redox reaction and electrochemical proton gradient can influence the number of protons pumped per reaction.
The Gibbs free energy associated with the redox chemistry carried out at complex I is described by eq. (1), which is simply the number of electrons transferred, multiplied by Faraday’s constant and the potential of the redox couple (E). The potential of the redox couple is described by the Nernst eq. (2), which is dependent on the standard state of the redox couple (E°), less the natural log of the ratio between the concentration of oxidized and reduced versions of each species.
| 1 |
| 2 |
Complex I homologs convert the free energy of these redox reactions into free energy stored in the proton transmembrane electrochemical gradient. This form of Gibbs free energy depends on the number of protons translocated (nH), multiplied by the difference in electrochemical proton potential across the membrane (eq. (3)). The electrochemical potential is in turn dependent on the pH and charge gradients across the membrane (eq. (4)).
| 3 |
| 4 |
Combining the two Gibbs free energy terms yields eq. (5), which gives the free energy change associated with the combined reactions of electron transfer and proton translocation.
| 5 |
Of particular interest for the discussion of these 2M complexes is the nH term. Solving eq. (5) for nH yields eq. (6) which illustrates how each of the variables affects nH.
| 6 |
Many different aspects of the reaction catalyzed by complex I homologs can affect the number of protons pumped, but only a few seem likely to require an increased proton pumping capacity during electron transport in the organisms we have identified that contain 2M complexes. For example, a change in the identity of electron carriers may result in an increase in E°: i.e., a larger redox potential difference between the cytoplasmic electron carrier and the membrane bound electron carrier. Due to the positive dependence of E° on nH in eq. (6), if all other factors remain the same, this change would increase the number of protons that can be pumped. Similarly, decreasing ΔpH will increase the value of nH, since there is an inverse relationship between these two values. Restating this in physiological terms: as the extracellular pH increases, more protons can be pumped per reaction since there is a weaker proton motive force resisting the action.
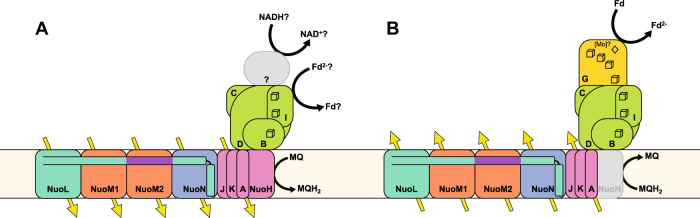
The effect ∆pH can have on enzymes relying on proton motive force has already received much attention in the context of alkaliphilic bacteria such as Bacillus pseudofirmus OF4. These organisms have adapted to high pH environments in part by increasing the number of c-ring subunits in their ATP synthases [39–42]. This adaptation increases the number of protons translocated per ADP phosphorylated, and helps compensate for the fact that each proton translocation carries with it less energy. We hypothesize that in some cases the addition of the second NuoM subunit in 2M complexes could be a complex I equivalent of increasing the number of c-ring subunits in the Bacillus ATPase (Fig. 7a). With respect to this hypothesis, it is worth noting that of the five Desulfovibrionales encoding 2M complexes, four are alkaliphiles with pH optima between 9.5 and 10 [43, 44].
Fig. 7.
Possible functions of 2M complexes. a Extra proton pumping subunit may be used to extrude a fifth proton per reaction in order to conserve energy under conditions of lowered transmembrane proton motive force, such as in alkaliphilic environments or potentially as an adaptation to slow growth. b The extra proton may be used for extra driving force to conduct reverse electron transport onto low potential electron acceptors like ferredoxin (Fd) for use in carbon or nitrogen fixation
In the case of alkaliphiles described above, the environment imposes a decrease in the electrochemical proton gradient. Another possible reason for an organism to operate at an electrochemical proton gradient below “normal” levels may be the adaptation to slow growth. Protons can leak directly through the cytoplasmic membrane in a way that is decoupled from energy conservation. For organisms that metabolize catabolic substrates very slowly, this leakage may present a large problem to their viability [45, 46]. The amount of proton leakage through a membrane is related to the electrochemical proton gradient across it [47]. In fact, the leakage current through the membrane appears to be non-linear with respect to transmembrane voltage, meaning a relatively small drop in transmembrane electrochemical proton gradient could result in a quite significant decrease in leakage current [47]. If the catabolic activity of a slow growing organism respiring at low rates cannot keep up with the leakage current experienced by fast-growing organisms at high respiration rates, then one possible solution would be to employ biochemical adaptations that decrease their normal operating transmembrane potential. Living at this lower potential would require the pumping of more protons to conserve the same amount of energy contained in a redox reaction. The addition of an additional M subunit in complex I may help accomplish this.
It is noteworthy that many of the organisms highlighted here, particularly Nitrospira and ammonia oxidizing Thaumarchaea, are notorious for slow growth, low biomass yields, and substrate limited growth conditions [48–51]. Additionally, three of the Verrucomicrobia (TAV1, TAV2 and TAV5) and Acidobacteria sp. KBS 89 were all isolated in a single study which described a novel cultivation strategy specifically designed for slow growing organisms [52]. Enrichments were incubated for >30 days and the colonies that were picked were invisible to the naked eye. Interestingly, the Verrucomicrobia were isolated from the termite gut, and the Acidobacteria from soil, but both were isolated under slow growth conditions and both contained a version of the 2M complex.
Finally, complex I can run in the reverse direction, producing low potential electron carriers (like NADH) from high-potential donors (like ubiquinonol) at the expense of proton motive force. This reverse electron transport occurs in many chemolithoautotrophs and anoxygenic photoautotrophs, which utilize electron donors that are at a higher redox potential than the cytoplasmic electron carrier required for carbon fixation [53]. The Nitrospira that comprise Clade 2 [51, 54] and the Thaumarchaea that comprise Clade 3 [35] both face this challenge with their autotrophic pathways. The ammonia-oxidizing Thaumarchaea utilize the highly efficient hydroxypropionate/hydroxybutyrate cycle for carbon fixation that requires electrons from NADPH [55], while Nitrospira utilize the reverse tricarboxylic acid cycle for carbon fixation, which requires the use of even lower potential ferredoxins [56]. In both cases reverse electron transport must occur, since the midpoint potential of NADPH (−320 mV) and ferredoxins (ca. −500 mV) are well below the potential of the NH4+/NO2− redox couple used for their energy metabolism (+340 mV) [57]. One hypothesis is that in these organisms, the 2M complexes are used specifically for reverse electron transport onto NADPH or ferredoxin (Fig. 7b). In Nitrospina gracilis it has been proposed that a complex I operon lacking a traditional N-module could be responsible for reverse electron transport to the level of ferredoxin [58]. Close relatives of that version of complex I do not appear in the Nitrospira genomes recovered to date, but the 2M complexes described here could in principal provide a similar energetic advantage under conditions of reverse electron transport. A similar hypothesis could explain the presence of 2M complexes in photosynthetic Chloroflexi as well. These organisms use reaction center II for cyclic electron flow, which, unlike reaction center I, does not produce a reduced ferredoxin [59]. Therefore, to withdraw electrons from the photosynthetic electron transport chain and transfer them to lower potential electron acceptors like NAD(P)H for carbon assimilation and anabolism, there must be an input of energy for reverse electron transport from quinols. It is possible that in these organisms reverse electron transport can only be achieved using the increased driving force of the 2M complexes.
Conclusions
Complex I and its homologous bioenergetic complexes have been modified and repurposed many times during the evolution of life on Earth. This evolution has involved the addition and removal of subunits specific for certain substrates onto a membrane-integral chassis containing between 1 and 3 large proton pumping subunits [20]. The 2M complex I homologs described here represent a fascinating modification on the canonical complex I enzyme that incorporate a fourth large pumping subunit. The phylogenetic analyses presented here demonstrate that these 2M complexes are polyphyletic, with respect to both the phylogeny of the host organism and the evolutionary history of the individual major pumping subunits. Combined with the unique gene synteny found in each of the three clades this strongly supports the conclusion that the construction of these operons resulted from multiple cases of convergent molecular evolution.
The highly conserved nature of the 2M complexes in each of the three respective clades implies that these are not pseudogenes or evolutionary dead-ends. The construction of gene clusters with the additional subunit, as well as the precise size and location of the insertions in the amphipathic helix of NuoL implies that this increase in pumping potential has been selected for under specific adaptive pressures. The apparent horizontal transfer of the second NuoM instead of a simple duplication may reflect the different environments in which the two proteins reside, one between NuoL and a NuoM, the other between a NuoM and NuoN. Perhaps finding a NuoM protein in sequence space that was able to function in this novel position was easier to achieve by sampling NuoM genes from other organisms instead of via duplication and modification of the original NuoM.
Future work is required to understand the conditions under which these complexes are expressed, and verify their subunit composition, substrate specificity, and proton pumping stoichiometry. However, the physiological diversity of the organisms containing these complexes suggests that they have evolved in response to different selective pressures and for different functions. The evolution of these complexes appears to have involved changes to the genetic content of an organism through horizontal gene transfer, genome restructuring through operon rearrangement, and protein primary, secondary, tertiary and quaternary structure modification in the extension of the NuoL amphipathic helix. Altogether, these observations highlight the different biological scales over which changes occur in order to bring about functional evolutionary transitions.
Electronic supplementary material
Acknowledgements
This publication was funded by the Gordon and Betty Moore Foundation through Grant #GBMF3780 and through work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research [DE - SC0016469] and the NASA Astrobiology Institute, award # NNA13AA92A (to V.J.O.). G.L.C. was supported by the NIH/NRSA training grant, T32 GM007616. We would like to thank Michael Wagner and Holger Daims for their thoughtful reading and comments on the manuscript, as well as the careful attention of an anonymous reviewer.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Grayson L. Chadwick, Phone: +805-451-4696, Email: chadwick@caltech.edu
Victoria J. Orphan, Phone: +626-395-1786, Email: vorphan@gps.caltech.edu
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0210-1) contains supplementary material, which is available to authorized users.
References
- 1.Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev. 1966;41:445–501. doi: 10.1111/j.1469-185X.1966.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 2.Bäumer S, Ide T, Jacobi C, Johann A, Gottschalk G, Deppenmeier U. The F420H2 dehydrogenase from methanosarcina mazei is a redox-driven proton pump closely related to NADH dehydrogenases. J Biol Chem. 2000;275:17968–73. doi: 10.1074/jbc.M000650200. [DOI] [PubMed] [Google Scholar]
- 3.Weerakoon DR, Olson JW. The campylobacter jejuni nadh:ubiquinone oxidoreductase (complex i) utilizes flavodoxin rather than NADH. J Bacteriol. 2008;190:915–25. doi: 10.1128/JB.01647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battchikova N, Eisenhut M, Aro EM. Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim Biophys Acta. 2011;1807:935–44. doi: 10.1016/j.bbabio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Welte C, Deppenmeier U. Membrane-bound electron transport in methanosaeta thermophila. J Bacteriol. 2011;193:2868–70. doi: 10.1128/JB.00162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–5. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 7.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–8. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329:448–51. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 9.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sazanov LA. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol. 2015;16:nrm3997. doi: 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- 11.Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A. The prokaryotic Mo/W-bisPGD enzymes family: A catalytic workhorse in bioenergetic. Biochim Biophys Acta. 2013;1827:1048–85. doi: 10.1016/j.bbabio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, et al. Comparative genomic analysis of the class epsilonproteobacteria and proposed reclassification to epsilonbacteraeota (phyl. nov.) Front Microbiol. 2017;8:682. doi: 10.3389/fmicb.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich T, Scheide D. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 2000;479:1–5. doi: 10.1016/S0014-5793(00)01867-6. [DOI] [PubMed] [Google Scholar]
- 14.Moparthi VK, Hägerhäll C. The evolution of respiratory chain complex I from a smaller last common ancestor consisting of 11 protein subunits. J Mol Evol. 2011;72:484–97. doi: 10.1007/s00239-011-9447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz O, Bothe H. The diaphorase subunit HoxU of the bidirectional hydrogenase as electron transferring protein in cyanobacterial respiration? Naturwissenschaften. 1996;83:525–7. doi: 10.1007/BF01141957. [DOI] [PubMed] [Google Scholar]
- 16.Gao F, Zhao J, Wang X, Qin S, Wei L, Ma W. NdhV is a subunit of NADPH dehydrogenase essential for cyclic electron transport in Synechocystis sp. Strain PCC 6803. Plant Physiol. 2016;170:752–60. doi: 10.1104/pp.15.01430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N, et al. The genome of the ammonia-oxidizing CandidatusNitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–45. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 18.Friedrich T, Steinmüller K, Weiss H. The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett. 1995;367:107–11. doi: 10.1016/0014-5793(95)00548-N. [DOI] [PubMed] [Google Scholar]
- 19.Jones AJY, Blaza JN, Varghese F, Hirst J. Respiratory complex I in Bos taurus and Paracoccus denitrificans pumps four protons across the membrane for every NADH oxidized. J Biol Chem. 2017;292:4987–95. doi: 10.1074/jbc.M116.771899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moparthi VK, Kumar B, Al-Eryani Y, Sperling E, Górecki K, Drakenberg T, et al. Functional role of the MrpA- and MrpD-homologous protein subunits in enzyme complexes evolutionary related to respiratory chain complex I. Biochim Biophys Acta. 2014;1837:178–85. doi: 10.1016/j.bbabio.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Belevich G, Knuuti J, Verkhovsky MI, Wikström M, Verkhovskaya M. Probing the mechanistic role of the long α-helix in subunit L of respiratory complex I from Escherichia coli by site-directed mutagenesis. Mol Microbiol. 2011;82:1086–95. doi: 10.1111/j.1365-2958.2011.07883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres-Bacete J, Sinha PK, Matsuno-Yagi A, Yagi T. Structural contribution of c-terminal segments of NuoL (ND5) and NuoM (ND4) subunits of complex I from Escherichia coli. J Biol Chem. 2011;286:34007–14. doi: 10.1074/jbc.M111.260968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deppenmeier U, Blaut M, Mahlmann A, Gottschalk G. Reduced coenzyme F420: heterodisulfide oxidoreductase, a proton- translocating redox system in methanogenic bacteria. Proc Natl Acad Sci USA. 1990;87:9449–53. doi: 10.1073/pnas.87.23.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welte C, Deppenmeier U. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta. 2014;1837:1130–47. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Markowitz VM, Chen IMA, Chu K, Szeto E, Palaniappan K, Jacob B, et al. IMG/M-HMP: a metagenome comparative analysis system for the human microbiome project. PLoS ONE. 2012;7:e40151. doi: 10.1371/journal.pone.0040151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talavera G, Castresana J, Kjer K, Page R, Sullivan J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 30.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE). 2010; 1–8.
- 31.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protocol. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20:714–37. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, López-García P. Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic thaumarchaeota and euryarchaeota. Genome Biol Evol. 2014;6:1549–63. doi: 10.1093/gbe/evu127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedrich T. The NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochim Biophys Acta. 1998;1364:134–46. doi: 10.1016/S0005-2728(98)00024-3. [DOI] [PubMed] [Google Scholar]
- 37.Efremov RG, Sazanov LA. The coupling mechanism of respiratory complex I — a structural and evolutionary perspective. Biochim Biophys Acta. 2012;1817:1785–95. doi: 10.1016/j.bbabio.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Cerqueira NMFSA, Gonzalez PJ, Fernandes PA, Moura JJG, Ramos MJ. Periplasmic nitrate reductase and formate dehydrogenase: similar molecular architectures with very different enzymatic activities. Acc Chem Res. 2015;48:2875–84. doi: 10.1021/acs.accounts.5b00333. [DOI] [PubMed] [Google Scholar]
- 39.Hicks DB, Liu J, Fujisawa M, Krulwich TA. F1F0-ATP synthases of alkaliphilic bacteria: lessons from their adaptations. Biochim Biophys Acta. 2010;1797:1362–77. doi: 10.1016/j.bbabio.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier T, Morgner N, Matthies D, Pogoryelov D, Keis S, Cook GM, et al. A tridecameric c ring of the adenosine triphosphate (ATP) synthase from the thermoalkaliphilic Bacillus sp. strain TA2.A1 facilitates ATP synthesis at low electrochemical proton potential. Mol Microbiol. 2007;65:1181–92. doi: 10.1111/j.1365-2958.2007.05857.x. [DOI] [PubMed] [Google Scholar]
- 41.Preiss L, Klyszejko AL, Hicks DB, Liu J, Fackelmayer OJ, Yildiz Ouml, et al. The c-ring stoichiometry of ATP synthase is adapted to cell physiological requirements of alkaliphilic Bacillus pseudofirmus OF4. Proc Natl Acad Sci USA. 2013;110:7874–9. doi: 10.1073/pnas.1303333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preiss L, Langer JD, Hicks DB, Liu J, Yildiz Ouml, Krulwich TA, et al. The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic lifestyle. Mol Microbiol. 2014;92:973–84. doi: 10.1111/mmi.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorokin DY, Tourova TP, Henstra AM, Stams AJM, Galinski EA, Muyzer G. Sulfidogenesis under extremely haloalkaline conditions by Desulfonatronospira thiodismutans gen. nov., sp. nov., and Desulfonatronospira delicata sp. nov. – a novel lineage of Deltaproteobacteria from hypersaline soda lakes. Microbiology. 2008;154:1444–53. doi: 10.1099/mic.0.2007/015628-0. [DOI] [PubMed] [Google Scholar]
- 44.Sorokin DY, Tourova TP, Kolganova TV, Detkova EN, Galinski EA, Muyzer G. Culturable diversity of lithotrophic haloalkaliphilic sulfate-reducing bacteria in soda lakes and the description of Desulfonatronumthioautotrophicum sp. nov., Desulfonatronum thiosulfatophilum sp. nov., Desulfonatronovibrio thiodismutans sp. nov., and Desulfonatronovibrio magnus sp. nov. Extremophiles. 2011;15:391–401. doi: 10.1007/s00792-011-0370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Niftrik AL, Fuerst JA, Damsté JSS, Kuenen JG, Jetten MSM, et al. The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol Lett. 2004;233:7–13. doi: 10.1016/j.femsle.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 46.Valentine DL. Opinion: adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 2007;5:nrmicro1619. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- 47.Rupprecht A, Sokolenko EA, Beck V, Ninnemann O, Jaburek M, Trimbuch T, et al. Role of the transmembrane potential in the membrane proton leak. Biophys J. 2010;98:1503–11. doi: 10.1016/j.bpj.2009.12.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium,Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 49.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–6. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 50.Stieglmeier M, Klingl A, Alves RJE, Rittmann SKMR, Melcher M, Leisch N, et al. Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol. 2014;64:2738–52. doi: 10.1099/ijs.0.063172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–9. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol. 2004;70:4748–55. doi: 10.1128/AEM.70.8.4748-4755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aleem MIH. Generation of reducing power in chemosynthesis II. Energy-linked reduction of pyridine nucleotides in the chemoautotroph, Nitrosomonas europaea. Biochim Biophys Acta. 1966;113:216–24. doi: 10.1016/S0926-6593(66)80062-0. [DOI] [PubMed] [Google Scholar]
- 54.Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci USA. 2015;112:11371–6. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Könneke M, Schubert DM, Brown PC, Hügler M, Standfest S, Schwander T, et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA. 2014;111:8239–44. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA. 2010;107:13479–84. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon J, Klotz MG. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim Biophys Acta. 2013;1827:114–35. doi: 10.1016/j.bbabio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol. 2013;4:27. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fischer WW, Hemp J, Johnson JE. Evolution of oxygenic photosynthesis. Annu Rev Earth Planet Sci. 2016;44:647–83. doi: 10.1146/annurev-earth-060313-054810. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.