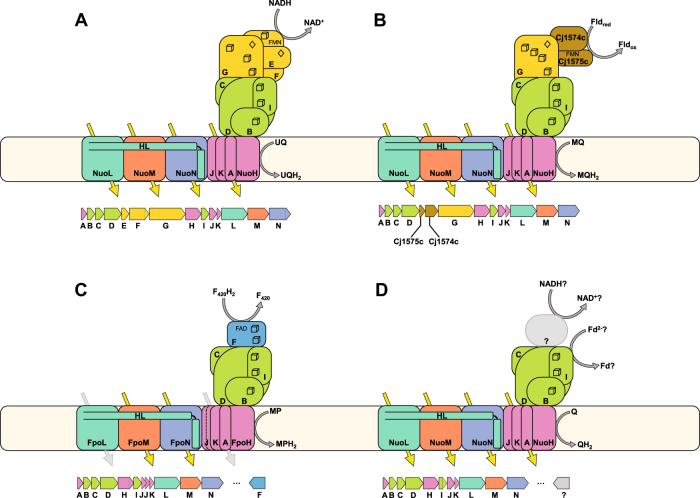
Fig. 1.
Common complex I homolog quaternary structure and gene order. Yellow arrows indicate proton translocations from the cytoplasm to the periplasm through the membrane-bound P-module. The C-terminal amphipathic helix of NuoL is labeled HL. 4Fe4S sulfur clusters are depicted as cubes, 2Fe2S clusters as diamonds. a Canonical 14 subunit NADH:ubiquinone oxidoreductase (Nuo) as found in T. thermophilus. Genes are generally arranged in an operon in alphabetical order as depicted. b A complex I cassette found in Camplylobacterota and best characterized in C. jejuni and H. pylori. The NuoEF genes are replaced by two smaller, non-homologous, proteins, which facilitate the interaction with flavodoxin (Fld) instead of NADH. The NuoG proteins in these operons contain an extra iron–sulfur cluster. c A methanogen-type F420H2:methanophenazine oxidoreductase (Fpo) showing the replacement of the N-module (NuoEFG) with the FpoF gene (in blue). This gene is not commonly associated with the rest of the operon, but its inclusion in the functional protein complex has been demonstrated by purification of the native Fpo complex. Note: two genes are annotated as NuoJ in these operons, but this represents a fission of the canonical NuoJ gene, and not a gene duplication event. d A common complex I gene cassette found in many organisms containing only the core 11 subunits of the complex. Few of these complexes have been studied biochemically and it is currently unknown whether the electron donors for these complexes are soluble proteinaceous electron carriers such as ferredoxins (Fd), or whether additional proteins encoded elsewhere in the genome are recruited to the complex to interact with small molecule electron carriers like NADH