Fig. 2.
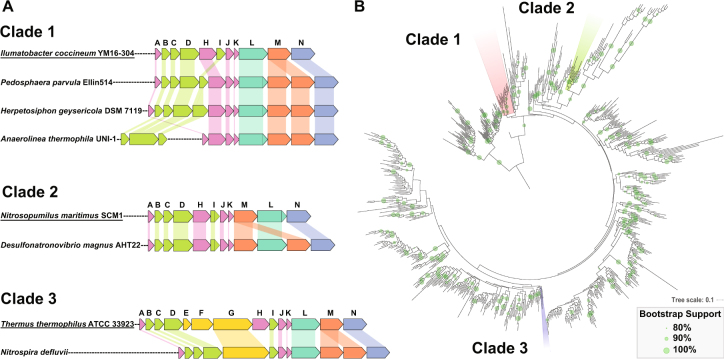
2M complex I homologs were found in three discrete clades. a Examples of the operon structure of 2M complexes from each of the three different clades showing the addition of a second NuoM subunit (in orange), as well as general operon features. Closely related canonical complex I operons are shown for comparison (underlined). In Clade 1 the second NuoM occurs right after the first, and the operons contain a unique gene rearrangement that swaps the NuoH and NuoI subunits; in some members of this Clade such as Anaerolinea thermophila UNI-1 the NuoBCDI genes occur separately from the rest of the operon. In Clade 2 the two NuoM genes are found on either side of NuoL; the NuoM preceding the NuoL is characteristic of the complex I operons found in Thaumarchaea. None of the operons have full N-modules, and only Clade 3 has a NuoG homolog in its operon. b A maximum likelihood phylogenetic tree of NuoL subunits from a rarified subset of publically available genome data. The NuoL genes from Clades 1, 2, and 3 fall far from one another in well-supported groups surrounded by NuoL genes in normal complex I operons—a pattern that implies that multiple independent events of convergent evolution gave rise to the 2M complex Is. Bootstrap support is shown for values ≥80%. High resolution NuoL trees, including those built from MUSCLE and MAFFT alignments, are shown in Figures S1-10