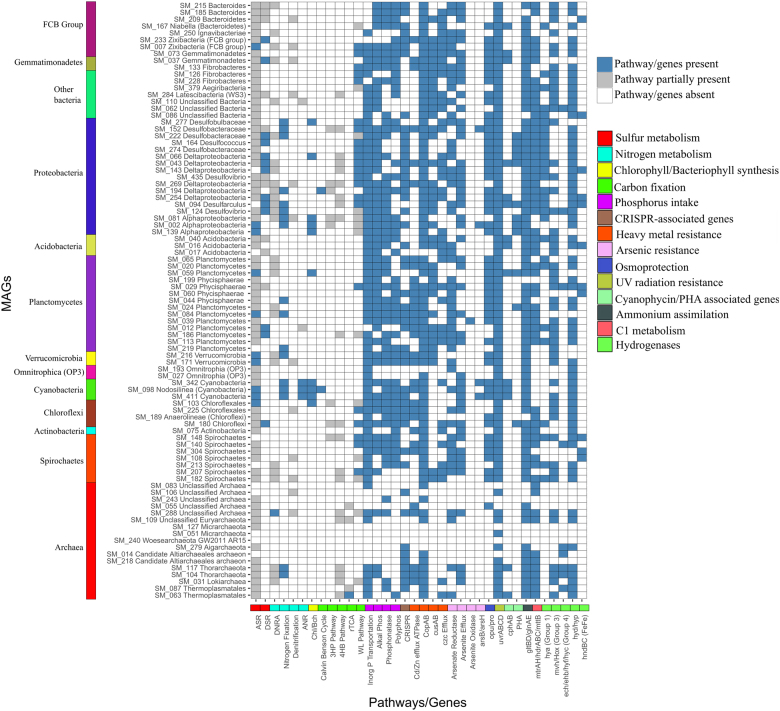
Fig. 5.
Color-coded table indicating major functional genes identified and their abundance in Shark Bay smooth mat draft MAGs. X-axis indicates specific genes likely involved in either nutrient cycling or environmental adaptation and y-axis the genome bin designations. Key: blue indicates the complete pathway identified in carbon, sulfur, and nitrogen nutrient cycles, genes present in heavy metal resistance, environmental adaptation, and hydrogenases; gray indicates the partial pathway identified in the carbon, sulfur, and nitrogen nutrient cycles; white indicates genes and pathways that are absent. On the left the color panel correspond to the phyla as shown in Fig. 1. On the right the colors represent different genes and pathways. Abbreviations, ASR assimilatory sulfate reduction, DSR dissimilatory sulfate reduction, ANR assimilatory nitrate reduction, Chl/Bch chlorophyll/bacteriochlorophyll production, 3HP pathway 3-hydroxypropinate pathway, 4HB pathway 4-hydroxybutyrate pathway, rTCAd reverse Krebs cycle, WL pathway Wood–Ljungdahl pathway (color figure online)