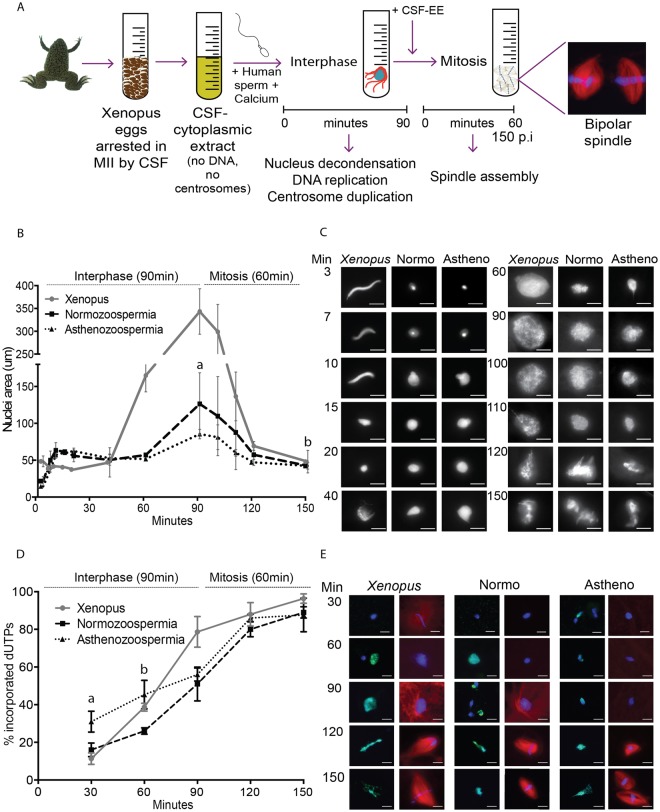
Figure 1.
Human spermatozoa nuclei reorganise and replicate their DNA in XEE. (A) Schematic representation of the experimental design. 3,000 human (or Xenopus) sperm nuclei and calcium were incubated in the XEE to mimic fertilization. These incorporations induce the resumption of the cell cycle, first by 90 min of interphase followed by 60 min of mitosis. (B,C) Time course of sperm nucleus decondensation. Xenopus and human spermatozoa (normozoospermic and asthenozoospermic) incubated in XEE were retrieved at different time points along the 150 min of the experiment, to measure the nuclei area. The graph shows the area evolution in µm2 of the three different sperm samples. aSignificantly different p-value of 0.010. bNo significantly different p-value of 0.89. Dispersion data are given as SD. The images on the right are representative pictures of the nuclei decondensation per time point. Scale bar, 10 µm. (D,E) Time course of sperm DNA replication. Xenopus and human (normozoospermic and asthenozoospermic) spermatozoa, calcium and biotin-labelled dUTPs were incubated in the XEE and retrieved every 30-min. The graph shows the percentage of incorporated dUTPs per sample. aSignificantly different p-value of 0.017. bSignificantly different p-value of 0.013. Dispersion data are given as Standard Deviation (SD). The images are representative pictures of the dUTPs incorporation. dUTPs are in green, DNA in blue and microtubules in red. Scale bar, 10 µm.