Figure 1. Overview and validation of VIVO.
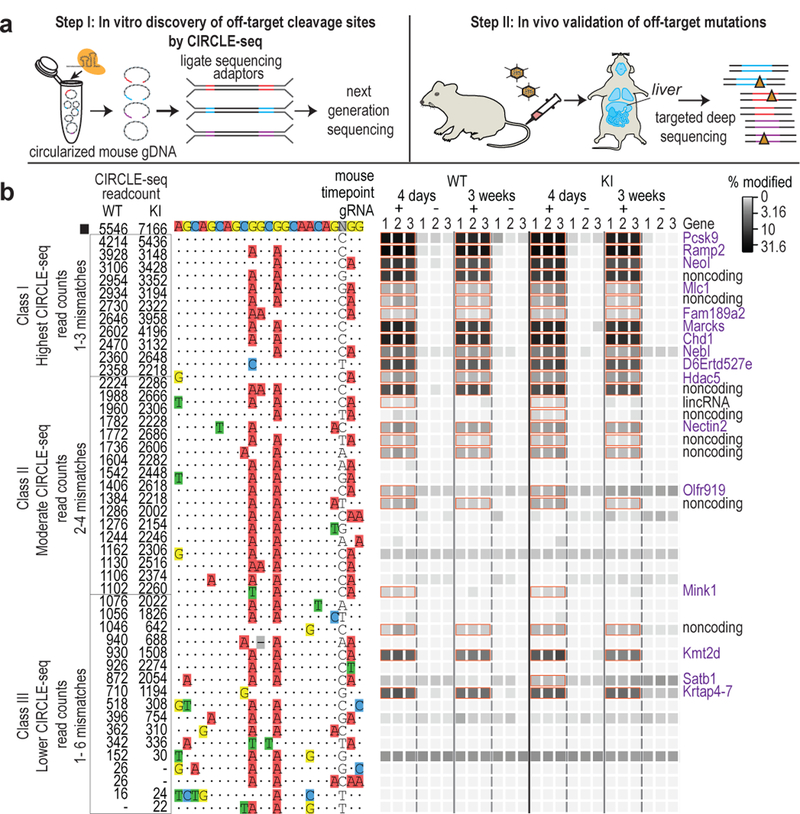
(a) Schematic illustrating the two-step VIVO method. In Step 1, CIRCLE-seq identifies off-target sites cleaved in vitro. In Step 2, the sites identified in Step 1 are assessed in vivo for indel mutations by targeted amplicon sequencing performed with genomic DNA isolated from the livers of nuclease-treated mice. (b) Assessment of in vivo off-target indels induced by gP/SpCas9. Indel frequencies as determined by targeted amplicon sequencing are presented as heat maps for the gP/SpCas9 on-target site (black square) and the Class I, Class II, and Class III off-target sites (identified from CIRCLE-seq experiments). Each locus was assayed in n=3 biologically independent WT and KI mice (1, 2, 3) using genomic DNA isolated from the liver of mice treated with experimental adenoviral vector encoding gP/SpCas9 (gRNA +) or control adenoviral vector GFP/SpCas9 (gRNA -). Mismatches relative to the on-target site are shown with colored boxes and spacer sequence is numbered from 1 (most PAM-proximal) to 20 (most PAM-distal). CIRCLE-seq read count numbers for each site are shown. Sites that showed a significant difference between the experimental gRNA + and gRNA - samples are outlined with orange boxes and labelled by genomic locus with coding regions shown in purple text. P values and significance were obtained by fitting a negative binomial generalized linear model (for source data and P values, see Supplementary Table 2).
