Figure 7.
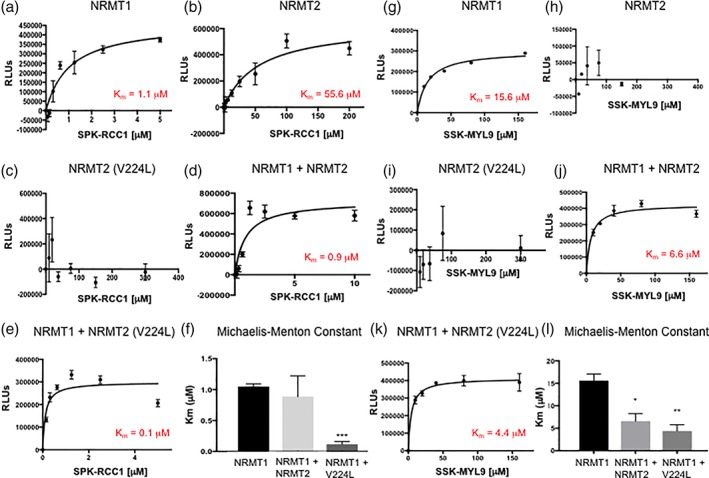
Catalytic activity of NRMT2 is not necessary for increased trimethylation by NRMT1. (a) Wildtype NRMT1 shows a K m of 1.1 μM when trimethylating the SPK‐RCC1 peptide substrate, (b) while the K m of NRMT2 is 55.6 μM. (c) The catalytically inactive NRMT2 V224L mutant is unable to methylate the same substrate. (d) Combining NRMT1 and NRMT2 lowers the K m of NRMT1 to 0.9 μM, while (e) the combination of NRMT1 and catalytically inactive NRMT2 V224L further lowers the K m of NRMT1 to 0.1 μM. (f) Comparison of Michaelis–Menton constants for NRMT1 alone, NRMT2 alone, or NRMT1 in combination with NRMT2 or NRMT2 V224L with SPK‐RCC1 peptide as substrate. Only the combination of NRMT1 with NRMT2 V224L was significantly different from NRMT1 alone. Calculated constants are the mean of three independent experiments ±SEM. *** denotes P < 0.001 as determined by unpaired Student's t‐test. Using a less preferred, but still enzymatically favorable substrate SSK‐MYL9, (g) increases the K m of NRMT1 to 15.6 μM but shows no enzymatic activity with (h) NRMT2 or (i) NRMT2 V224L. (j) Combining NRMT1 and NRMT2 decreases the K m of NRMT1 to 6.6 μM, and (k) the combination of NRMT1 and NRMT2 V224L further lowers the K m of NRMT1 to 4.4 μM. (l) Comparison of Michaelis–Menton constants for NRMT1 alone, NRMT2 alone, or NRMT1 in combination with NRMT2 or NRMT2 V224L with SSK‐MYL9 peptide as substrate. The constants for both the combinations of NRMT1 with NRMT2 and NRMT1 with NRMT2 V224L were significantly different from NRMT1 alone. Calculated constants are the mean of three independent experiments ±SEM. * and ** denote P < 0.05 and 0.01, respectively, as determined by unpaired Student's t‐test.
