Fig. 3.
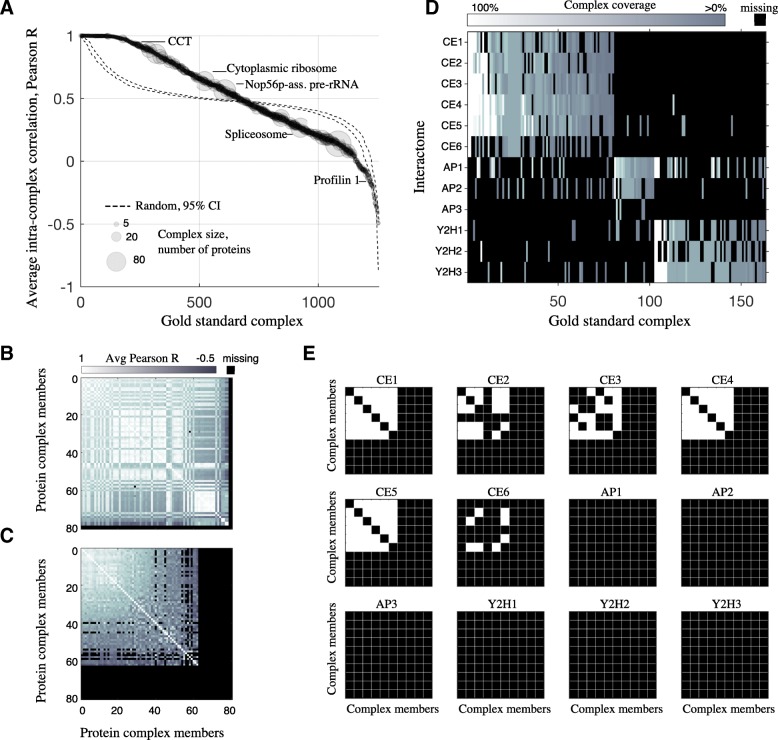
High-throughput techniques consistently recover some gold standard complexes and consistently fail to recover others. a Average internal, pairwise correlation for every quantified gold standard complex. Only gold standard complexes with at least two identified proteins in one of 20 co-fractionation datasets are shown (1253/2652 CORUM complexes). Correlation values are pooled across the 20 co-fractionation datasets, and the number of internal, pairwise comparisons is given by marker size. The pattern expected by random chance is shown (dashed lines, 95% CI). b Connection matrix, cytoplasmic ribosome. Pairwise correlation values were averaged over 20 datasets. Proteins ordered by average pairwise correlation. c C complex spliceosome. d Technique-specific gold standard complexes. All gold standard complexes predicted by at least 2/3 of the published interactomes from a given technique (CF, AP, Y2H), and no more than 1 interactome from the other techniques. e Connection matrices for the Chaperonin Containing TCP-1 complex (CCT), a gold standard complex, taken from the published interactomes. White: interacting protein pair. Black: non-interacting