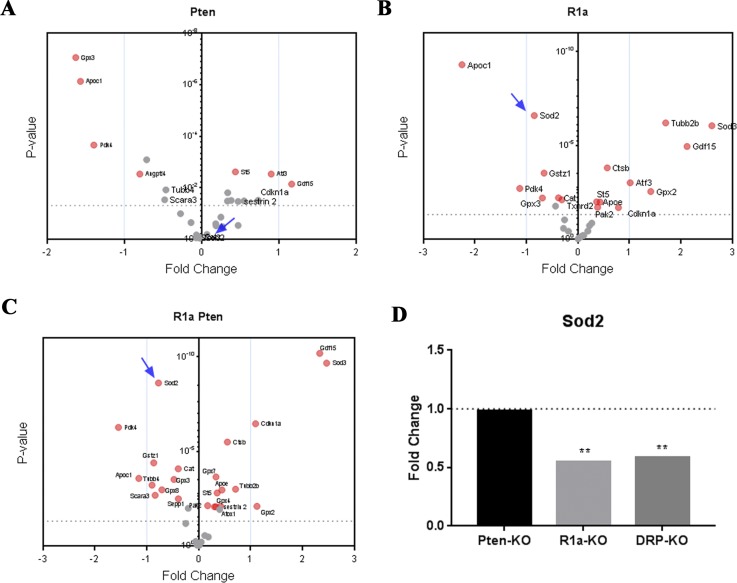
Figure 2.
Deregulation of OS genes in murine tumor progression models. Microarray data comparing Pten-, R1a-, and DRP-TpoKO tumors. Oxidative pathways genes were analyzed in (A) Pten-TpoKO tumors, (B) R1a-TpoKO tumors, and (C) DRP-TpoKO tumors. Significantly altered genes above cutoff (P < 0.01) are indicated by red circles; those below the cutoff are indicated by gray circles; Sod2 is indicated by the arrow in panels A–C. Wild-type Cre-negative littermates were used for comparison for each model. (D) Expression of Sod2 in each tumor model compared with wild-type control. Dotted line represents the fold change of Cre-negative control tissue for each group. **P < 0.01.