 1,3,4-Thiadiazole compounds containing catechol moiety and chalcone motif are synthesized and examined for antioxidant activity, cytotoxicity and DNA-binding activity.
1,3,4-Thiadiazole compounds containing catechol moiety and chalcone motif are synthesized and examined for antioxidant activity, cytotoxicity and DNA-binding activity.
Abstract
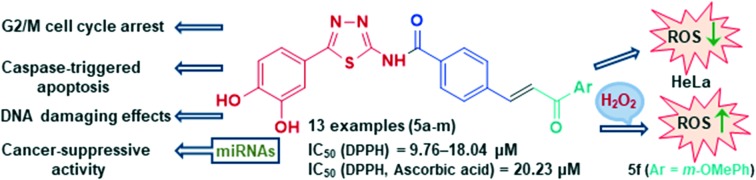
Hybrid compounds that combine the 1,3,4-thiadiazole-containing catechol moiety with a chalcone motif were synthesized and examined for their antioxidant activity, cytotoxicity, and DNA-binding activity. A series of thirteen compounds showed strong antioxidant and cytotoxic effects on human acute promyelocytic leukemia HL-60 cells. Several compounds exerted good cytotoxic activities on cervical adenocarcinoma HeLa cells. The treatment of HeLa cells with IC50 and double IC50 concentrations of the compounds 5a, 5c, 5f, and 5m induced a statistically significant increase in the percentage of cells within a subG1 cell cycle phase. The examined compounds caused G2/M cell cycle arrest in HeLa cells. Each of these compounds triggered apoptosis in HeLa cells through activation of caspase-3, the main effector caspase, caspase-8, which is involved in the extrinsic apoptotic pathway, and caspase-9, which is involved in the intrinsic apoptotic pathway. All of the examined compounds decreased the expression levels of MMP2 in HeLa cells and levels of protumorigenic miR-133b. Compounds 5a and 5m lowered the expression level of oncogenic miR-21 in HeLa cells. In addition, compounds 5a, 5f, and 5m decreased the expression levels of oncogenic miR-155 while the treatment of HeLa cells with compounds 5a, 5c, and 5f increased expression of tumor-suppressive miR-206. Observed effects of these compounds on expression levels of four examined miRNAs suggest their prominent cancer-suppressive activity. An investigation by absorption and fluorescence spectroscopy showed more efficient calf thymus DNA binding activity of the compound 5m in comparison to other tested compounds. Results of a pUC19 plasmid cleavage study and comet assay showed DNA damaging activities of compounds 5a and 5c.
1. Introduction
Anticancer hybrid molecules incorporate two or more different covalently linked pharmacophores with ability to modulate multiple biological targets and improve therapeutic potential of designed compounds in comparison to single bioactive precursors. Using molecular hybridization techniques, it is possible to synthesize numerous hybrids based on known anticancer scaffolds leading to a more favorable pharmacological profile than the sum of each individual compound.1 Chalcones are frequently selected as one of several privilege structures because of their significant anticancer properties and facile preparation, offering major advancements in the field of hybrid molecules.2,3 Such chalcone hybrid analogues with promising anticancer activity have already been synthesized by the coupling of chalcones with various bioactive compounds including coumarin,4,5 1,2,3-triazole,6 retinoid,7 naphthoquinone,8 β-carboline,9 artemisinin,10N-4-piperazinyl-ciprofloxacin,11 anthraquinone,12 pyrazole-5-carboxamide,13 thiazole,14 and isoxazole.15
We selected 1,3,4-thiadiazole as a second bioactive compound to combine with a chalcone unit to generate a new hybrid molecule suitable for simultaneously targeting several pathogenic mechanisms. A covalent bond was established by formation of an amide in the reaction of the amine group of the heterocycle and a previously transformed carboxylic function into an acid chloride of the chalcone analogue. 1,3,4-Thiadiazoles exhibited a wide spectrum of anticancer activities owing to high electron-donating ability of nitrogen atoms to form hydrogen bonds or to coordinate metal ions.16,17 Although 1,3,4-thiadiazoles have a promising anticancer potential, their toxicity still remains a major concern.18 Molecular hybridization may be a way to enhance activity or selectivity and also overcome the side effects associated with the single compound.
It is well known that antioxidants (including phenolic ones) block free radicals that induce damage in biological macromolecules under oxidative stress, thus preventing their oxidation, DNA mutations, and malignant changes.19 Very recently, we combined bioactive functions of 1,3,4-thiadiazole and a phenolic acid moiety to obtain novel conjugates with antioxidant and antiproliferative activity.20 The reported antiproliferative activities of 1,3,4-thiadiazoles and chalcone analogues, as well as radical scavenging properties of phenolic compounds, led us to a rational design and synthesis of new molecular hybrids with evaluation of their antioxidant, cytotoxic, and DNA-binding potential.
2. Results and discussion
2.1. Chemistry
The novel 1,3,4-thiadiazole–chalcone hybrids were synthesized in three steps (Scheme 1). Initially, chalcone analogues 4a–m were prepared using the Claisen–Schmidt condensation of 4-formylbenzoic acid 3 with various substituted acetophenones in the presence of NaOH, according to a slightly modified literature procedure.21 Subsequently, chalcones 4a–m were converted into acyl chlorides by action of SOCl2 and, without isolation, reacted with 4-(5-amino-1,3,4-thiadiazol-2-yl)benzene-1,2-diol 2 in dioxane giving the final hybrid compounds 5a–m in moderate to good yields (53–87%). Previously, a thiadiazole derivative was synthesized by the reaction of 3,4-dihydroxybenzoic acid and a thiosemicarbazide in phosphoryl chloride.20 Although acid chlorides are very reactive compounds, the formation of an amide requires long time and high temperature due to poor nucleophilicity of the amino group of 1,3,4-thiadiazole. Almost all compounds still contained a significant amount of dioxane. The solvent was eliminated by dissolving crude product in DMF (dimethylformamide) or DMSO (dimethyl sulfoxide) and subsequent precipitation with water or by complete evaporation of THF (tetrahydrofuran) solutions. Structures of all compounds were confirmed by means of 1H and 13C NMR spectroscopy (see Supplementary content), IR, and elemental analysis. The olefinic protons of the chalcone double bond in hybrid compounds 5a–m, as well as their corresponding precursors 4a–m, appeared as an AB system. On the basis of coupling constant values (J = 15.6–16.0 Hz), the compounds 5a, 5c, 5d, 5f, 5g, and5i–m were isolated and characterized in E-isomeric form. However, compounds 5b, 5e, and 5h, and their chalcone precursors (4b, 4e, and 4h) having their coupling constant values in a range from 7.0 to 8.2 Hz, correspond to the less stable Z-configuration.22 This kind of geometry of the chalcone double bond is not very frequent and since these derivatives contain an ortho-substituted acetophenone moiety in their structure, it is probable that the Z-isomeric form is favored due to steric factors. Aromatic protons corresponding to the phenolic group appeared in the range of 6.86–7.44 ppm, giving a doublet for H-5, doublet of doublets for H-6, and another doublet for H-2. Protons belonging to the benzoic acid moiety were well resolved in the form of the AB system for most of the thiadiazol–chalcone derivatives.
Scheme 1. Reagents and conditions: a) POCl3, rt, H2NHNC( S)NH2, 1 h, reflux; b) substituted acetophenones, NaOH, MeOH, 2 h, reflux, HCl; c) SOCl2, DMF, CH2Cl2, 2 h, rt; d) 2, dioxane, 12 h, reflux.
2.2. Biology
2.2.1. Radical scavenging activity and effects of pretreatment of HeLa cells with 1,3,4-thiadiazole–chalcone hybrids on ROS levels
Antioxidant activity of phenolic acids is well known because of their preventive effect on malignant changes that are associated with radical species.23 Reactive oxygen species (ROS) in the form of free radicals (superoxide and hydroxyl radical) and neutral molecules (H2O2) have been neutralized by stable phenolic antioxidants, diminishing DNA damaging and cancer formation.19 Thus, compounds showing antioxidant and cytotoxic activity have a great importance and a combination of phenolic antioxidant moiety with other bioactive pharmacophores could be a good way to obtain more potent radical scavengers as a result of their synergistic effects.
Generally, all synthesized hybrid molecules exhibited better DPPH radical scavenging activity than referent ascorbic acid (Table 1).
Table 1. DPPH scavenging activity of the 1,3,4-thiadiazole–chalcone hybrids 5a–m a .
| Compound | IC50 ± SD (μM) |
| 5a | 12.03 ± 0.50 |
| 5b | 11.66 ± 0.41 |
| 5c | 17.17 ± 0.38 |
| 5d | 11.81 ± 0.31 |
| 5e | 9.76 ± 0.75 |
| 5f | 10.33 ± 0.30 |
| 5g | 17.86 ± 0.03 |
| 5h | 13.91 ± 0.32 |
| 5i | 17.82 ± 0.65 |
| 5j | 18.04 ± 0.23 |
| 5k | 14.01 ± 0.34 |
| 5l | 13.45 ± 0.09 |
| 5m | 11.06 ± 0.24 |
| Ascorbic acid | 20.23 ± 0.14 |
aResults are mean values ± SD from three measurements.
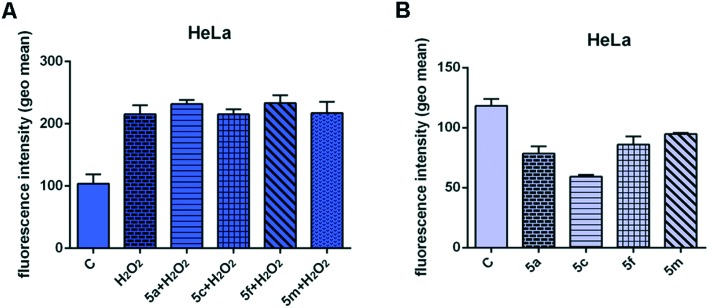
Unfortunately, all our compounds were practically insoluble in methanol, a common solvent for DPPH tests, so determination of radical scavenging activity was performed in diluted DMSO. However, DMSO was found to considerably decrease antiradical capacity of ascorbic acid due to the formation of a molecular complex with DMSO through intermolecular hydrogen bonds.24 Also, dramatic solvent effects were established on the rates of H-abstraction from phenolic compounds.25 In accordance with these facts, in separate experiments we observed significantly lower scavenging activity at higher concentrations of DMSO, suggesting the formation of a molecular complex between DMSO and our compounds. For this reason we were not able to consider the influence of the chalcone moiety with various electron donating or withdrawing substituents and make a comparison with other 1,3,4-thiazole compounds containing phenolic hydroxyl groups. Anyway, the potent radical scavenging potential of our compounds is evident, and for that reason we decided to explore their effects on ROS levels in human cervical adenocarcinoma HeLa cells. We examined the effects of four compounds – 5a, 5c, 5f, and 5m – which were selected for all further analyses due to their prominent cytotoxic activity and good selectivity in antiproliferative actions (Table 2). As can be seen in Fig. 1A, significant accumulation of ROS in the H2O2-treated cells indicates activation of endogenous ROS production by added H2O2. Pretreatment of HeLa cells with subtoxic IC20 concentrations of 5a and 5f for 24 h (10 μM for each compound) slightly increased the ROS levels induced by H2O2, while pretreatment with compounds 5c and 5m did not affect the ROS levels triggered by H2O2. Radical species (phenoxyl radical, etc.) produced by the oxidation of 5a and 5f with H2O2 are highly reactive and subject to further oxidation giving quinone compounds. Quinones are still reactive and can be stabilized by interaction with a nucleic acid.26 This reaction might be responsible for the toxic pro-oxidant effect of 5a and 5f which may damage cellular constituents like DNA by induction of oxidative cytotoxic stress in cancer cells, activating programmed cell death – apoptosis.
Table 2. Cytotoxic activity of the investigated 1,3,4-thiadiazole–chalcone hybrids containing an antioxidant phenolic moiety a .
| Compd. | IC50 ± SD (μM) |
|||
| HeLa | HL-60 | A549 | MRC-5 | |
| 5a | 9.37 ± 0.86 | 8.40 ± 1.44 | 42.75 ± 2.77 | 36.00 ± 1.10 |
| 5b | 9.12 ± 1.19 | 7.62 ± 1.40 | 21.80 ± 2.55 | 18.56 ± 2.25 |
| 5c | 9.63 ± 1.26 | 8.39 ± 1.39 | 27.87 ± 4.05 | 34.25 ± 4.88 |
| 5d | 11.20 ± 2.32 | 8.44 ± 1.52 | 26.35 ± 1.90 | 33.72 ± 3.35 |
| 5e | 10.22 ± 0.73 | 11.97 ± 1.27 | 50.23 ± 5.35 | 48.01 ± 6.43 |
| 5f | 9.98 ± 0.99 | 9.92 ± 0.50 | 92.14 ± 6.64 | 42.54 ± 3.88 |
| 5g | 57.55 ± 7.58 | 16.35 ± 0.87 | 89.10 ± 9.35 | 81.33 ± 9.25 |
| 5h | 11.08 ± 2.10 | 8.88 ± 1.32 | 46.89 ± 1.71 | 39.58 ± 4.82 |
| 5i | 17.75 ± 0.17 | 10.63 ± 0.96 | 89.33 ± 2.99 | 45.81 ± 3.38 |
| 5j | 40.22 ± 3.64 | 9.74 ± 1.54 | 87.74 ± 6 .82 | 38.24 ± 4.95 |
| 5k | 54.76 ± 2.77 | 11.17 ± 0.52 | 127.75 ± 8.37 | 39.44 ± 3.49 |
| 5l | 11.04 ± 2.41 | 6.92 ± 0.53 | 23.23 ± 1.37 | 21.17 ± 1.23 |
| 5m | 12.72 ± 3.24 | 15.72 ± 1.66 | 47.85 ± 1.92 | 68.52 ± 7.69 |
| Cisplatin | 4.91 ± 0.74 | 2.88 ± 0.34 | 13.21 ± 0.89 | 9.35 ± 1.29 |
aResults are mean values ± SD of three independent experiments.
Fig. 1. Effects of 24 h pretreatment of HeLa cells with IC20 concentrations of the 1,3,4-thiadiazole–chalcone hybrids 5a, 5c, 5f, and 5m (10 μM for each compound) on ROS generation induced by hydrogen peroxide (10 mM) (A) and endogenous ROS levels (B). The results are presented as the mean ± SD of two independent experiments.
In the absence of exogenously added H2O2, the intracellular ROS levels in HeLa cells treated for 24 h with each of the four compounds were decreased when compared with basal ROS levels in the control. This behavior of all tested compounds, especially 5c, is a result of their radical scavenging potential to neutralize basal ROS in HeLa cells (Fig. 1B). Some cancer cells, in advanced stage of disease, have adapted to oxidative stress due to their antioxidative defense capacity. Therefore, the cancer cells show resistance to drugs that induce intracellular ROS production, such as paclitaxel or doxorubicin.27 Thus, the possibility of drugs to reduce an antioxidant system in combination with oxidant agents might be useful in antitumor therapies. A redox modulation of the ROS level by production of a sufficient amount of ROS provoking apoptosis, or by decrease in antioxidant level, could be a good way to kill cancer cells which are more sensitive to exogenous oxidative stress than normal cells.
2.2.2. Cytotoxic activity
Cytotoxicity of 1,3,4-thiadiazole–chalcone hybrids containing an antioxidant catechol moiety was evaluated against three human malignant cell lines (cervical carcinoma HeLa, acute promyelocytic leukemia HL-60, and lung carcinoma A549) and normal human lung fibroblasts MRC-5 using the MTT cell survival test. The obtained IC50 values are shown in Table 2. Generally, all tested compounds exerted the strongest cytotoxic activity against leukemia HL-60 cells with IC50 values in a range from 6.92 μM to 16.35 μM. Compounds 5a, 5f, 5h, 5l, and 5m also showed strong cytotoxic effects on HeLa cells, with IC50 values from 9.12 μM to 12.72 μM. Lung carcinoma A549 cells were the least sensitive to the cytotoxic activity of the examined hybrids, while other tested derivatives exerted moderate to low cytotoxic activity. As seen in Table 2, there is no significant difference between the influence of electron-donating and electron-withdrawing groups of the acetophenone moiety on the cytotoxic action against cancer cells. This indicates that the thiadiazole–chalcone pharmacophore present in the structure of the tested compounds has the crucial role in their antiproliferative action. All of the compounds from this series exerted two to four times higher cytotoxic activity against HeLa and HL-60 malignant cell lines in comparison to their activity against normal MRC-5 cells, with the exception of 5g, 5j, and 5k against HeLa cells. In addition, all tested derivatives showed lower toxicity against normal MRC-5 cells compared to cisplatin as a referent chemotherapeutic.
2.2.3. Effects of the 1,3,4-thiadiazole–chalcone hybrids on cell cycle phase distribution
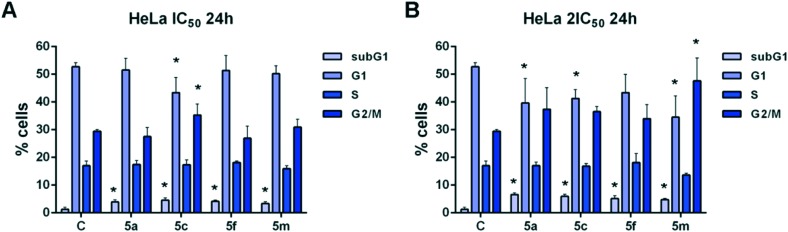
With the aim to investigate the mechanisms of the cytotoxic activity of four selected 1,3,4-thiadiazole–chalcone hybrids 5a, 5c, 5f, and 5m, changes in cell cycle phase distribution of cervical adenocarcinoma HeLa cells treated with IC50 and 2IC50 concentrations of these compounds for 24 h were assessed. Incubation with both tested concentrations of the compounds induced statistically significant increases in the percentage of HeLa cells within the subG1 phase when compared with untreated control cell samples (Fig. 2). These results point to ability of the examined compounds to induce cell death in HeLa cells. Furthermore, compound 5c applied at IC50 concentration and compound 5m applied at double IC50 concentration caused significant increases in the percentage of cells within a G2/M cell cycle phase in comparison to those percentages in the control cells. The incubation of HeLa cells with double IC50 concentrations of compounds 5a, 5c, and 5f also led to accumulation of cells in the G2/M phase, although those differences were not statistically significant. The obtained data demonstrated that tested compounds cause G2/M cell cycle arrest in HeLa cells, thus preventing cell entry into mitosis and eventually leading to cell apoptosis. The double IC50 concentrations of compounds 5a and 5m, as well as IC50 and double IC50 concentrations of compound 5c, induced significant decreases in the percentage of treated cells within the G1 phase when compared with control cells, suggesting enhanced sensitivity of cells within a G1 phase to the cytotoxic action of these compounds. Our findings are in accordance with literature data showing that compounds bearing a chalcone bioactive scaffold induce G2/M cell cycle arrest and apoptotic cell death in cancer cells.28
Fig. 2. Changes in the cell cycle phase distribution of human cervical carcinoma HeLa cells treated with IC50 (A) and 2IC50 concentrations (B) of the 1,3,4-thiadiazole–chalcone hybrids 5a, 5c, 5f, and 5m after 24 h treatment. Results are presented as the mean ± SD of three independent experiments. Statistically significant differences between control and treated cell samples are marked with *(p < 0.05).
2.2.4. Effects of the specific caspase inhibitors
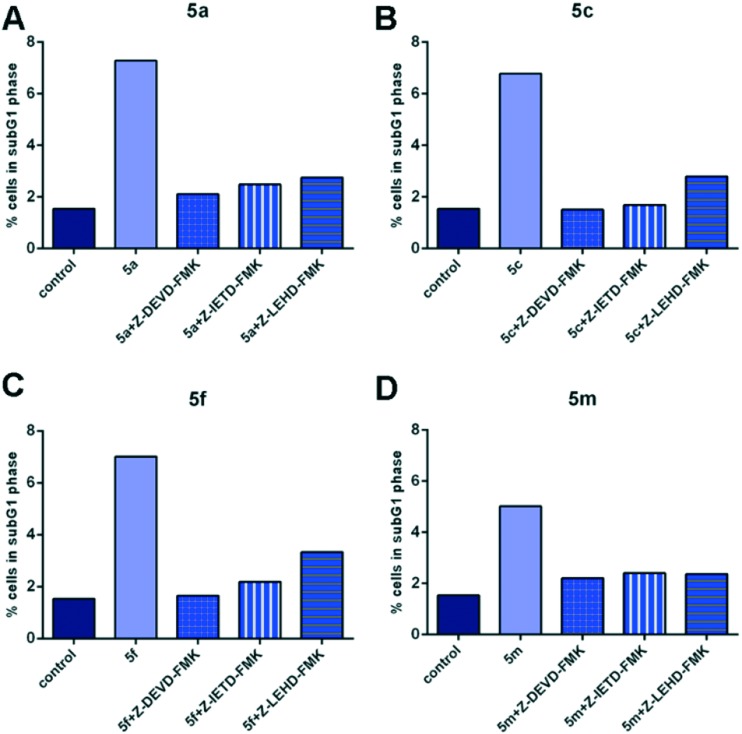
To further examine whether the tested compounds could induce apoptotic cell death in treated HeLa cells, the cells were pretreated with specific peptide inhibitors of caspase-3, caspase-8, or caspase-9 two hours before addition of compounds and their effects were determined by cell cycle analysis. As seen in Fig. 3, a notable decrease in the percentage of HeLa cells within the subG1 cell cycle phase was observed in cell samples which were pretreated with caspase-3 inhibitor, caspase-8 inhibitor, or caspase-9 inhibitor, and afterwards exposed to compounds 5a, 5c, 5f, or 5m when compared with HeLa cell samples which were not pretreated with inhibitors before addition of a test compound. The obtained results indicate that examined 1,3,4-thiadiazole–chalcone hybrids (5a, 5c, 5f, and 5m) trigger apoptosis in HeLa cells through the activation of main effector caspase-3, activation of caspase-8 implicated in extrinsic pathway of apoptosis, and caspase-9 implicated in intrinsic pathway of apoptosis.
Fig. 3. Effects of the specific caspase inhibitors (Z-DEVD-FMK – caspase-3 inhibitor, Z-IETD-FMK – caspase-8 inhibitor, Z-LEHD-FMK – caspase-9 inhibitor) on the percentages of HeLa cells within subG1 phase treated with 2IC50 concentrations of the compounds 5a (A), 5c (B), 5f (C), and 5m (D). Representative graphs are shown.
2.2.5. Effects of 1,3,4-thiadiazole–chalcone hybrids on gene and microRNA expression levels
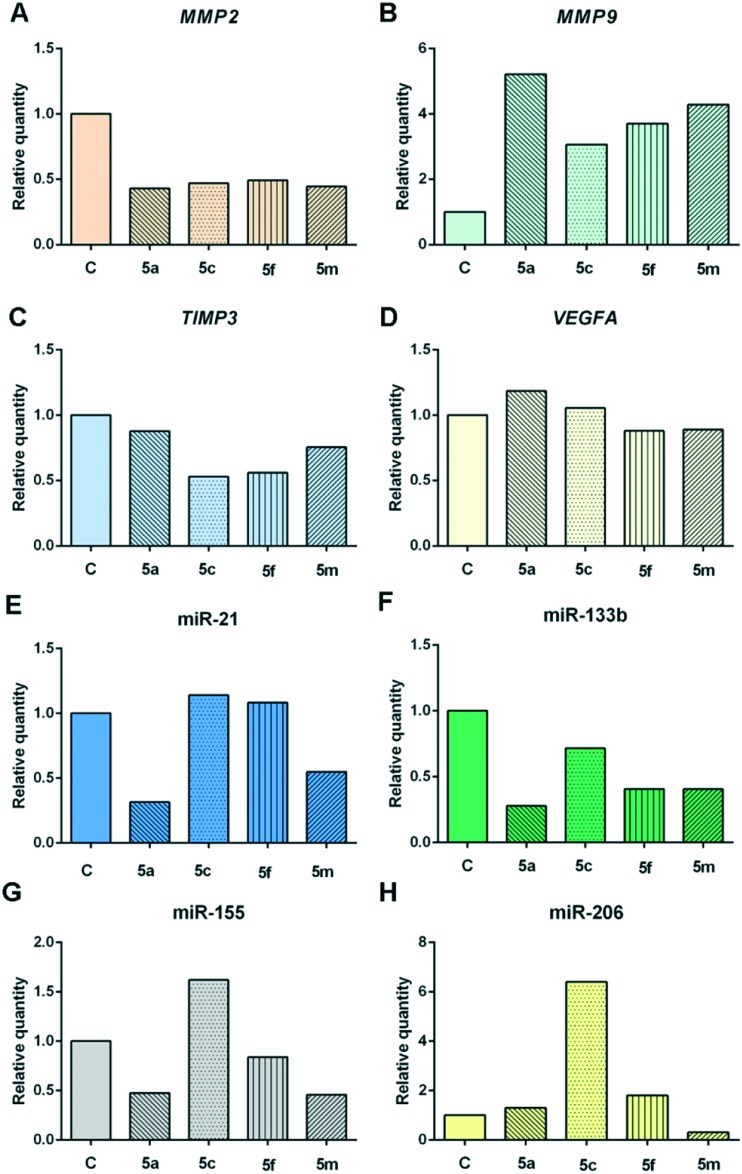
Increased levels of matrix-metalloproteinase-2 (MMP2), matrix-metalloproteinase-9 (MMP9), and vascular endothelial growth factor A (VEGFA) and increased levels of typical oncogenic miRNAs (miR-21 and miR-155) overexpressed in various cancer types,29 and decreased levels of matrix-metalloproteinase inhibitor 3 (TIMP3)30 are associated with cell growth, migration, epithelial mesenchymal transition, invasion, metastasis, and angiogenesis.31 MiR-133b is known to be downregulated in prostate cancer, but this microRNA exerted protumorigenic activity in cervical cancer.32 MiR-206 is described to be downregulated in cervical cancer,33 and that overexpression induces apoptosis.34
To further elucidate the molecular mechanisms of cytotoxic activity of our compounds, we examined changes in the expression levels of extracellular matrix-degrading proteases MMP2 and MMP9, TIMP3 which inhibits matrix metalloproteinases and regulates proteolysis of extracellular matrix, as well as in the expression of inducer of angiogenesis VEGFA, and several microRNAs-miR-21/133b/155/206 in human cervical adenocarcinoma HeLa cells treated with subtoxic IC20 concentrations of the examined compounds (Fig. 4).
Fig. 4. Changes in expression levels of MMP2 gene (A), MMP9 gene (B), TIMP3 gene (C), VEGFA gene (D), miR-21 (E), miR-133b (F), miR-155 (G), and miR-206 (H) in HeLa cells exposed to subtoxic IC20 concentrations of the compounds 5a, 5c, 5f, and 5m (10 μM for each compound) for 24 h. Representative graphs are shown.
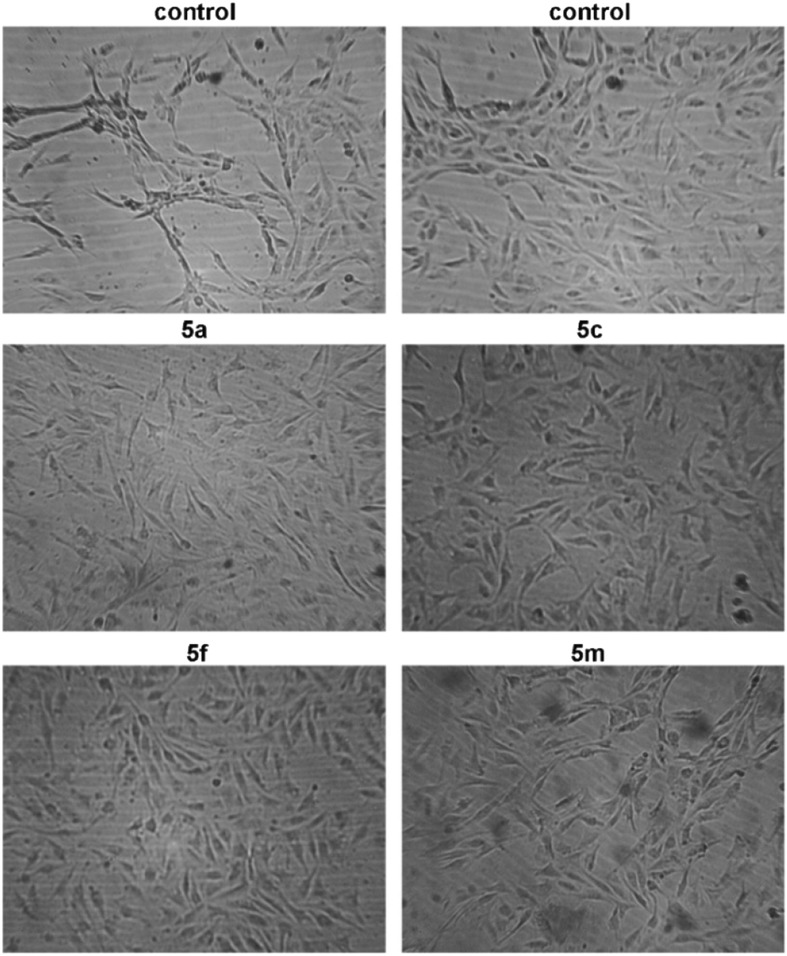
All of the examined compounds (5a, 5c, 5f, and 5m) decreased expression levels of MMP2 and TIMP3, but increased expression levels of MMP9, when compared with those levels in untreated, control HeLa cells. In addition, compound 5a slightly increased the VEGFA expression level in HeLa cells, while compounds 5f and 5m slightly decreased the expression of VEGFA in these cells. The observed downregulation of VEGFA expression in HeLa cells and inhibition of formation of tubular structures by EA.hy926 cells exerted by compounds 5f and 5m indicate the mild antiangiogenic activities of these compounds (Fig. 5).
Fig. 5. Photomicrographs of control EA.hy926 cells and EA.hy926 cells incubated with subtoxic IC20 concentrations of the compounds 5a, 5c, 5f (6.5 μM for each compound), and 5m (8 μM) for 20 h.
The ability of the examined 1,3,4-thiadiazole–chalcone hybrids to downregulate expression levels of MMP2 in HeLa cells might suggest their suppressive effects on cervical cancer cell invasion and establishment of metastasis. The investigation by Kato et al. suggested that expression levels of MMP2 may be closely associated with invasion ability of cervical cancer cells.35 We observed that all tested compounds remarkably increased MMP9 expression levels in HeLa cells. MMP9 is connected with increased invasion and metastasis, but besides this protumorigenic role, there is experimental evidence that MMP9 may exert protective effects on cancer progression and metastasis in addition to an anti-angiogenic effect.36,37 A study in a mouse model of multistage skin carcinogenesis caused by HPV16 oncogene demonstrated that MMP9 deficient transgenic mice had decreased proliferation rate of keratinocytes and a lowered incidence of invasive tumors, which had a more aggressive phenotype, while mice expressing MMP9 developed a larger number of tumors, which had a less aggressive phenotype.37 Furthermore, MMP9 may suppress further progression of malignant tumors through generation of various anti-angiogenic peptides, such as angiostatin and tumstatin.36 Taken together, these findings might suggest that upregulation of MMP9 expression levels in tumor stroma which could be induced by our compounds might contribute to suppression of tumor growth and progression.
Considering the effects of these compounds on microRNA expression levels in HeLa cells, compound 5a decreased levels of three crucial oncogenic miRNAs-miR-21/155 and protumorigenic miR-133b in cervical cancer, while it slightly increased level of tumor suppressive miR-206, when compared with those levels measured in control cells. Compound 5c remarkably increased expression levels of miR-206 and miR-155 in comparison to control HeLa cells, and decreased expression of protumorigenic miR-133b. Treatment with compounds 5c and 5f caused mild increase in miR-21 expression in HeLa cells. Exposure to compound 5f caused decrease in the expression of miR-133b and miR-155 in addition to an increase in the expression level of miR-206. Compound 5m lowered the expression levels of all four examined miRNAs when compared with controls cells. Our research showed that compounds 5a and 5m induced remarkable decrease in the expression level of oncogenic miR-21, that could result in reduced proliferation, migration, and invasion of cervical carcinoma cells. These tumor-suppressive effects had been shown to be regulated through phosphatase and a tensin homolog (PTEN).38 The antiproliferative potential of our four compounds is further proved by their ability to reduce levels of oncogenic miR-133b, which had been reported to be involved in enhanced proliferation and colony formation of cervical cancer cells, as well as in further progression and metastasis of cervical cancers, affecting AKT1 and ERK signaling pathways.39 Moreover, compounds 5a, 5f, and 5m lowered the expression levels of miR-155, which is overexpressed in cervical cancer tissue, stimulates cell proliferation, and exerts oncogenic activity, confirming their cancer-suppressive properties.40 The pro-apoptotic effects of compounds 5a, 5c, and 5f could be, at least in part, attributed to the increased expression of tumor-suppressor miR-206 in treated HeLa cells. The miR-206 had been reported as an inducer of apoptosis in HeLa cells, which was associated with inhibition of neurogenic locus notch homolog protein 3 (Notch 3).34
The investigated compounds and miRNAs changed the examined gene expression levels. TIMP3 is a proven target of miR-21, MMP9 is a target of miR-133b, while VEGF mRNA is target of miR-206 according to TargetScan Human 7.0 (; http://www.targetscan.org/).41
2.2.6. DNA binding study
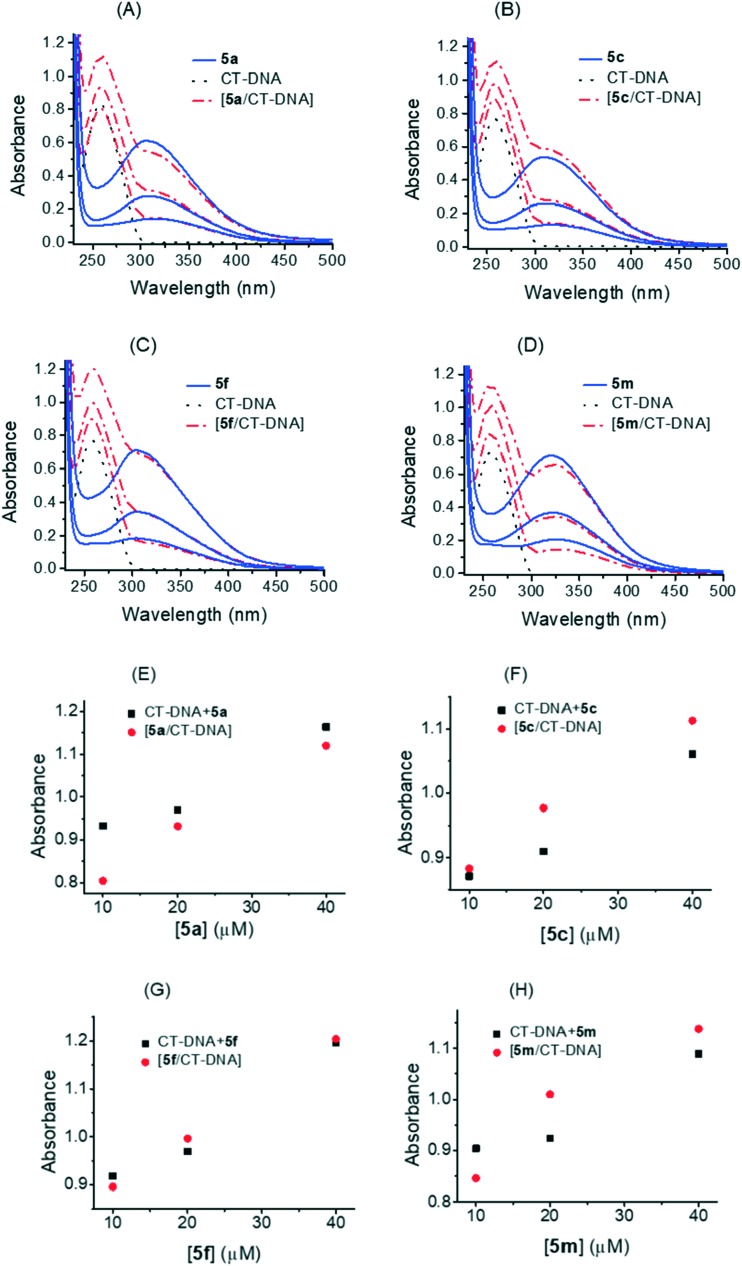
As is well known, the interaction of DNA with a small molecule can give rise to changes in the absorbance and in the peak position in an absorption spectra. Hyperchromism and hypochromism are regarded as spectral evidence for DNA double-helix structural change when DNA reacts with other molecules. Hyperchromism originates from the disruption of the DNA duplex secondary structure and is indicative of partial or non-intercalative modes42 and hypochromism originates from the stabilization of the DNA duplex by either the intercalation binding mode or the electrostatic effect of small molecules.43,44 Electronic absorption spectra of biologically most active 1,3,4-thiadiazole–chalcone hybrids recorded at different concentrations without or with fixed concentration of CT-DNA are shown in Fig. 6A. UV-vis spectra of all compounds displayed similar absorption bands. It was found that the maximum absorption of 5a, 5c, 5f, and 5m was centered at 309 nm, 310 nm, 305 nm and 323 nm, respectively. Upon interaction with CT-DNA, the formation of a compound–CT-DNA occured with no shift of absorption maximum at 259 nm. Detailed absorption changes induced by binding of the compounds to CT-DNA were further calculated from experimental data, Fig. 6B. The value of the sum of absorbances at 259 nm of a free compound and free CT-DNA was a different from the absorption value of a compound–CT-DNA.
Fig. 6. A. Changes in UV-vis absorption spectra of CT-DNA (95 μM) after interaction with different concentrations of 1,3,4-thiadiazole–chalcone hybrids. Panels A–D: UV-vis absorption spectra of 5a (10 μM, 20 μM, and 40 μM), 5c (10 μM, 20 μM, and 40 μM), 5f (10 μM, 20 μM, and 40 μM), and 5m (10 μM, 20 μM, and 40 μM), respectively before and after interaction with CT-DNA. B. Changes in UV-vis absorption spectra of CT-DNA (95 μM) after interaction with different concentrations of 1,3,4-thiadiazole–chalcone hybrids. Panels E–H: comparison of absorption at 259 nm between the CT-DNA–1,3,4-thiadiazole–chalcone compounds and the sum values of CT-DNA and 1,3,4-thiadiazole–chalcone compounds; representative absorption assay curves are shown. Triplicate assays were applied to all compounds and control CT-DNA.
A hypochromism of about 13% was observed with lower concentrations of 5a, Fig. 6B (panel E), while a decrease in absorption intensity at 259 nm was less pronounced at higher concentrations. In the case of 5c, Fig. 6B (panel F), hyperchromism was observed (–13.7%, –7.36%, and –4.9% at 10 μM, 20 μM, and 40 μM, respectively) and with 5m, Fig. 6B (panel H), hyperchromism was also observed, probably more pronounced at lower concentrations. The DNA showed the least changes after interaction with 5f, Fig. 6B (panel G), (this weak hypochromism was calculated as 2.4% at the lowest concentration of the compound). Previous studies demonstrated that a significant hypochromic effect with a concomitant red shift upon ligand binding to DNA was the typical characteristic of the intercalating mode.45,46 In this work, the obtained hypochromic effects of 5a and 5f and hyperchromic effects of 5c and 5m with no red shift probably reflect changes in the conformation and structure of CT-DNA upon binding of the compounds in the minor groove, via formation of hydrogen bonds between hydroxyl groups of 1,3,4-thiadiazole–chalcone parts of the compounds and exposed AT base pairs.
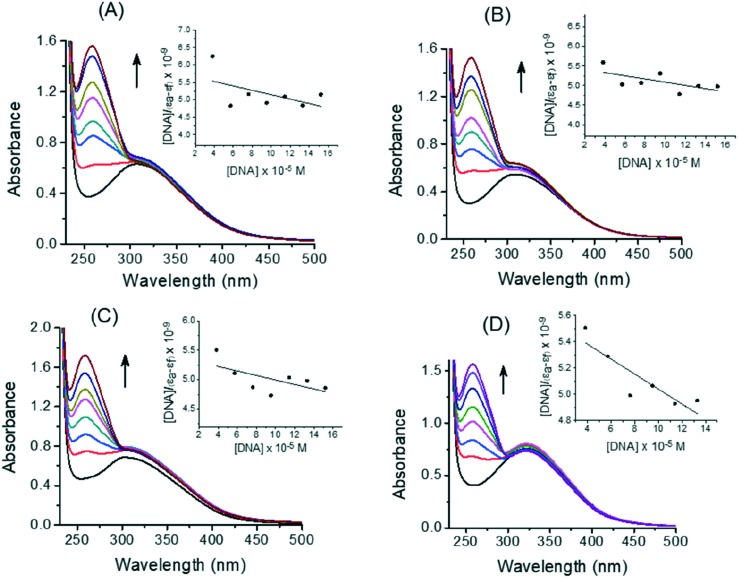
Stability of binding between CT-DNA and 1,3,4-thiadiazole–chalcone compounds was determined by a spectroscopic titration. Absorption spectra of 5a, 5c, 5f, and 5m without and with CT-DNA at different concentrations are shown in Fig. 7, panels A–D. Absorbance at 259 nm was monitored for each concentration of DNA (insets in Fig. 7 show plots after linearization). Binding constant Kb of 5a, 5c, 5f, and 5m were calculated using eqn (2) and values of 1.09 × 103 M–1, 0.74 × 103 M–1, 0.71 × 103 M–1, and 1.01 × 103 M–1, respectively, were obtained. These values are comparable with ones previously published for anthraquinone–chalcone hybrids12 and other structurally different compounds.47,48 However, relatively low correlation coefficients, indicate low binding strength of the interaction of 1,3,4-thiadiazole–chalcone hybrids with the helix of CT-DNA.
Fig. 7. Representative absorption titration curves of 5a (A), 5c (B), 5f (C), and 5m (D) at fixed concentration (40 μM) with increasing concentrations of CT-DNA (2.2, 4.4, 6.6, 8.8, 11, and 13 × 10–5 M). Insets: Determination of binding constant (Kb) by plot of [DNA]/(εa – εf) vs. [DNA]; the arrows show the changes in absorbance upon increasing amounts of CT-DNA. Triplicate assays were applied to all compounds and control CT-DNA.
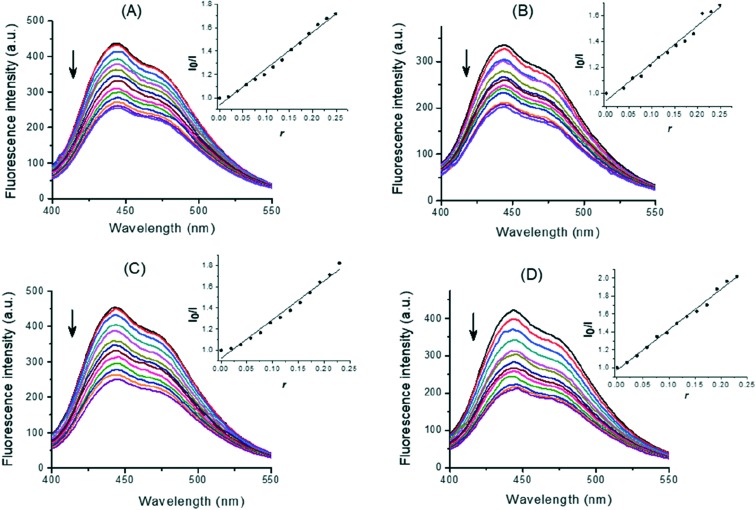
In order to provide additional insight into the interactions between the DNA and 1,3,4-thiadiazole–chalcone hybrids, a study with the minor groove binder Hoechst 33258 was performed. Hoechst 33258 (H) binds strongly and selectively with high affinity to the double-stranded B-DNA structure and, like other minor groove binders, it recognizes at least four AT base pairs. It binds by a combination of hydrogen bonding, van der Waals contacts with the walls of the minor groove, and electrostatic interactions between its cationic structure and the DNA.49 Binding of Hoechst 33258 to CT-DNA was followed by excitation at 350 nm with a maximum in fluorescence at 444 nm. The fluorescence intensity of the band at 444 nm of the Hoechst–CT-DNA system decreased remarkably with increasing concentration of the compounds (Fig. 8). The fluorescence intensities of H–CT-DNA–compound systems were 58%, 60%, 55%, and 51% of a H–CT-DNA system at maximal applied concentrations of 5a, 5c, 5f, and 5m, respectively. The observed reduction of fluorescence indicated the propensity of the 1,3,4-thiadiazole–chalcone hybrids to bind to DNA minor groove. Insets in Fig. 8 show the quenching plots demonstrating that quenching of H bound to CT-DNA by these compounds is in agreement with the linear Stern–Volmer eqn (3) for the investigated concentration ranges of the compounds. The corresponding quenching constants of H–CT-DNA system for 5a, 5c, 5f, and 5m were calculated by linear regression of the plot I0/I versus [compound]/[CT-DNA] as K = 3.30, 2.95, 3.95, and 4.66, respectively.
Fig. 8. Displacement of DNA-bound Hoechst 33258 (H) by 5a (A), 5c (B), 5f (C), and 5m (D). Emission spectra (λex = 350 nm) of H (2.8 × 10–5 M) bound to CT-DNA (1 × 10–4 M, top line) and quenching of H–CT-DNA system by the compounds at increasing concentrations (0–2.2 × 10–5 M, curves from top to bottom). Insets: Fluorescence curves of EB bound to CT-DNA at λmax = 444 nm by 1,3,4-thiadiazole–chalcone compounds; r = [compound]/[CT-DNA]. The arrow shows that fluorescence intensity decreased with increasing concentration of the complex; representative fluorescence assay curves are shown. Triplicate assays were applied to all compounds and control CT-DNA.
Comparing K values for the quenching fluorescence intensity obtained by the displacement experiments, it may be concluded that 5m with a thiophene ring was more efficient as a groove binder than other tested 1,3,4-thiadiazole–chalcone hybrids with benzene rings. However, considering the extent of the interaction of the compounds with CT-DNA, it can be concluded that these compounds bind to DNA exhibiting minor groove mode with low binding strength, Also, the results suggested that the differences in a benzoic moiety contribute to binding, as well as the binding in the groove most likely occurs via forming hydrogen bonds among hydroxyl groups of 1,3,4-thiadiazole–chalcone parts of the compounds and exposed AT base pairs.
2.2.7. Comet assay
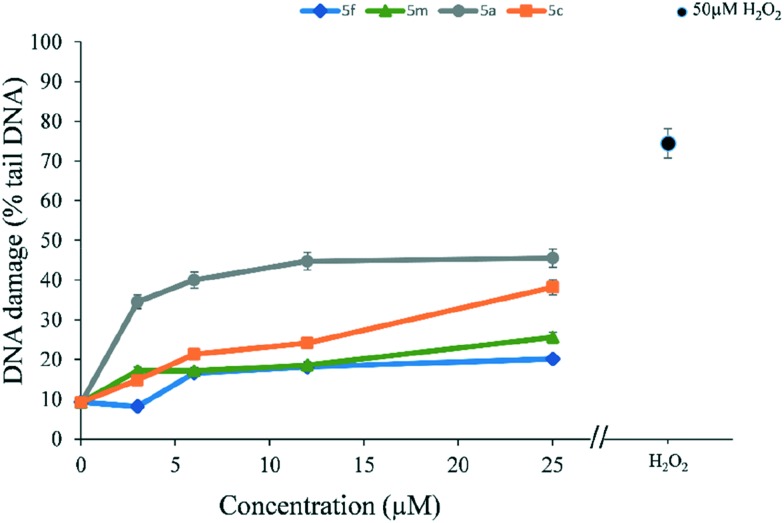
Comet assay50 is a versatile and sensitive method for measuring DNA damage in terms of single and double-strand breaks in DNA. In order to evaluate the extent of DNA damage induced by selected compounds a comet assay was performed on MRC-5 cells which were treated for 24 h with different concentration of compounds 5a, 5c, 5f, and 5m. From the results presented in Fig. 9 it can be seen that the compounds caused DNA damage, evidenced by an increase in% of DNA in the comet tail. Each of the tested compounds exhibited concentration-dependent DNA damage in MRC-5 cells. At the highest concentration tested (25 μM), DNA damage detected was 45% in case of 5a compound, while in the case of 5c DNA damage was 38%. Lower degrees of DNA damage were detected in case of compounds 5m and 5f: 25% and 20%, respectively.
Fig. 9. Detection of DNA damage (% tail DNA) using the Comet assay with MRC-5 cells after 24 h exposure with compounds 5f, 5m, 5a, and 5c (3.125–25 μM). PBS was used as negative control, and H2O2 (50 μM, 5 min in-gel exposure) was used as positive control. The y-axis shows the mean ± SD of DNA damage measured through tail intensity parameter. The experiments were conducted in triplicate and repeated twice.
2.2.8. pUC19 DNA cleavage study
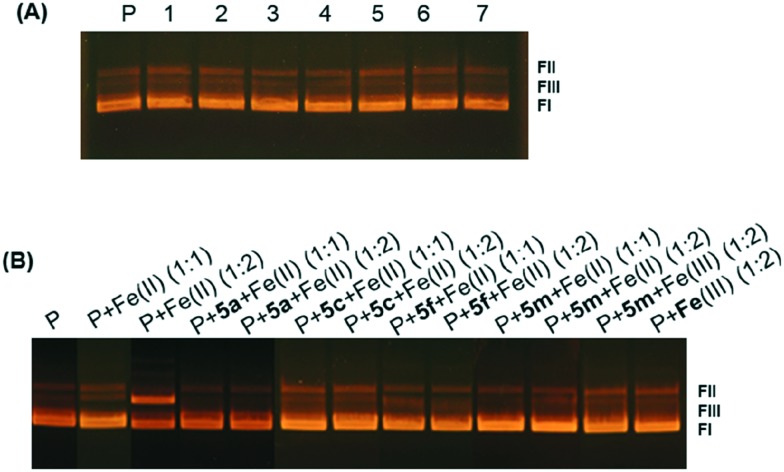
Since a weak DNA damaging activity was detected in cell cultures, the abilities of most potent 1,3,4-thiadiazole–chalcone hybrids to cleave double-stranded plasmid DNA were investigated using an agarose electrophoretic assay and DMSO as solvent for compounds in the performed concentration range (from 0.5–20%) which had no effects on conformation of plasmid DNA (results not shown). As shown by a representative agarose gel electrophoresis in Fig. 10(A), lane 1, control plasmid pUC19 consisted mainly of supercoiled form FI and nicked form FII, with also a small quantity of linear form FIII. All of the compounds were tested, and the results showed that there were no significant changes in band mobility, and no strand scission under the applied range of concentrations. In Fig. 10(A) results obtained with compound 5a are shown. Very similar results were obtained with compounds 5c, 5f, and 5m (not shown). Further, strand scission was tested under reducing conditions, in presence of iron(ii). Iron(ii) per se is involved in the damage of DNA51 and the results of cleavage of pUC19 by iron(ii) is shown in Fig. 10(B), lanes P + Fe(ii) (1 : 1) and P + Fe(ii) (1 : 2). When the compound and iron were used in mole ratio 1 : 1, (Fig. 10(B), lanes P + 5a + Fe(ii) (1 : 1), P + 5cFe(ii) (1 : 1), P + 5f + Fe(ii) (1 : 1), and P + 5m + Fe(ii) (1 : 1) and mole ratio 1 : 2 ((Fig. 10(B), lanes P + 5a + Fe(ii) (1 : 2), P + 5cFe(ii) (1 : 2), P + 5f + Fe(ii) (1 : 2), and P + 5m + Fe(ii) (1 : 2), 1,3,4-thiadiazole–chalcone hybrids abolished the effects of iron, probably by complex formation and/or by anti-oxidative action of the catechol moiety. Iron Fe(iii) had no effect under any conditions, Fig. 10(B), lanes P + 5m + Fe(iii) (1 : 2) and P + Fe(iii) (1 : 2). These results indicate that DNA cleavage could not be the cause of cytotoxicity. The obtained protective activities of 1,3,4-thiadiazole–chalcone hybrids are comparable to those published for anthraquinone–chalcone hybrids.12
Fig. 10. Representative agarose gel electrophoresis of plasmid pUC19 treated with 1,3,4-thiadiazole–chalcone hybrid: (A) plasmid pUC19 (12 nM) without (lane P) and with 5a at concentration of 0.05 mM, 0.1 mM, 0.15 mM, 0.2 mM, 0.25 mM, 0.3 mM, and 0.4 mM, lanes 1, 2, 3, 4, 5, 6, and 7, respectively; (B) effects of 5a, 5c, 5f, and 5m on Fe(ii) induced pUC19 damage with compound-iron mole ratios 1 : 1 and 1 : 2.
3. Conclusion
Combining two pharmacophores, 1,3,4-thiadiazole derived from protocatechuic acid with antioxidant properties and a series of chalcones, thirteen novel hybrid compounds were prepared and evaluated for radical scavenging, cytotoxic, and DNA-binding potential. The synthesized 1,3,4-thiadiazole–chalcone hybrids containing a phenolic moiety exerted a strong antioxidant and moderate to good cytotoxic activity against HL-60 and HeLa cells. The selected compounds 5a, 5c, 5f, and 5m caused G2/M cell cycle arrest in HeLa cells and showed the ability to trigger apoptosis in HeLa cells through activation of caspase-3, caspase-8, and caspase-9. All examined compounds remarkably downregulated the expression levels of MMP2 in treated HeLa cells, suggesting their suppressive effects on cervical cancer cell invasion and metastasis. Compounds 5a and 5m induced a remarkable decrease in the expression levels of oncogenic miR-21 in HeLa cells while all four examined compounds lowered levels of protumorigenic miR-133b. In addition, compounds 5a, 5f, and 5m decreased the expression levels of oncogenic miR-155. Treatment of HeLa cells with compounds 5a, 5c, and 5f elevated expression of tumor-suppressor miR-206. These effects of the test compounds on expression levels of four examined miRNAs point to their prominent cancer-suppressive properties. 1,3,4-Thiadiazole–chalcone hybrids bind weakly to the helix of CT-DNA in the minor groove, induce weak DNA damage in cell culture, and protect plasmid DNA from iron(ii)-induced DNA damage. But, DNA displacement of fluorescent probes and plasmid DNA damage activity as well as damage DNA and comet assay indicated their DNA damaging potential. Taken together, the results of our research indicate promising antiproliferative properties of four newly synthesized 1,3,4-thiadiazole–chalcone hybrids containing phenolic moiety.
4. Experimental section
4.1. Chemistry
4.1.1. Physical measurements and methods
Melting points were determined on a Mel-Temp capillary melting points apparatus, model 1001 and are uncorrected. Elemental (C, H, N, S) analysis of the samples was carried out in the Center for Instrumental Analysis, Faculty of Chemistry, Belgrade. UV spectra were recorded using an Agilent Technologies, Cary 300 Series UV-vis spectrophotometer. IR spectra were obtained on a PerkinElmer Spectrum One FT-IR spectrophotometer with a KBr disc. 1H and 13C-NMR spectra were recorded on a Varian Gemini 200 MHz spectrometer.
4.1.2. Procedure for the preparation of 5a–m
Procedure for the synthesis of 2
A mixture of 3,4-dihydroxybenzoic acid, 1 (2.00 mmol, 0.308 g,) and POCl3 (1.0 mL) was stirred for 20 minutes at room temperature. Then, thiosemicarbazide (2.50 mmol, 0.228 g) was added and the resulting suspension was refluxed for 1 hour. After cooling the flask in an ice bath, 3.0 mL of distilled water was carefully added, and reflux was continued for 1 hour. The mixture was then cooled to room temperature, a saturated aqueous solution of NaOH was added until pH 8.5 was reached, and the suspension was stirred for 1 hour at room temperature. The formed precipitate of the corresponding 1,3,4-thiadiazole derivative (2) was then filtered, dried over CaCl2, and recrystallized from hot 50% aqueous EtOH.
Procedure for the synthesis of 4
A mixture of 4-formylbenzoic acid (2.3 mmol, 0.345 g), a corresponding substituted acetophenone (2.00 mmol) and solid NaOH (4.6 mmol, 0.184 g) in dry methanol (10.0 mL) was refluxed for 2 hours. After completion, the mixture was cooled to room temperature, 5.0 mL of distilled water was added, and pH of the solution was adjusted to 4.5 with HCl aqueous solution (2 M), upon which a precipitate was formed. The precipitate (4a–m) was stirred for 30 minutes and then subsequently collected by suction filtration, dried over CaCl2, and recrystallized from hot MeOH.
Procedure for the synthesis of 5a–m
To the mixture of 4 (1.00 mmol) in dry dichloromethane (4.0 mL), SOCl2 (4.00 mmol, 0.3 mL) was slowly added, followed by two drops of dimethylformamide. The resulting mixture was then stirred for 2 h at room temperature. Afterwards, the solvent was evaporated under reduced pressure, and the excess of SOCl2 was removed by azeotropic distillation with toluene. 2-Amino-1,3,4-thiadiazole, 2 (1.00 mmol, 0.227 g) and dry dioxane were added to the formed acid chloride, without its previous isolation, and the mixture was then refluxed for 12 hours. After cooling, 20.0 mL of distilled water was slowly added to the solution with vigorous stirring at room temperature, followed with a formation of a precipitate (5a–m). Stirring was continued for 1 hour and the precipitate was then filtrated, washed with water, and dried over CaCl2. All compounds (except 5e) contained a smaller quantity of dioxane which was not removed even after drying at 110 °C for 12 h. To remove dioxane, final compounds 5j and 5l were dissolved in N,N-dimethylformamide and 5k was dissolved in dimethyl sulfoxide, after which distilled water was added, followed with formation of a precipitate. The formed precipitate was then filtrated, washed with water, and dried over CaCl2. All other final compounds (5a–d, 5f–i, and 5m) were dissolved in tetrahydrofuran and the solvent was evaporated to dryness under reduced pressure.
4.1.2.1. (E)-4-(3-Oxo-3-phenylprop-1-en-1-yl)benzoic acid (4a)
1H NMR (200 MHz, DMSO-d6): 7.55–7.69, (m, 3H, Ar–H); 7.78, (d, 1H, JAB = 15.8 Hz, CH ); 8.00, (s, 4H, Ar–H); 8.06, (d, 1H, JBA = 15.8 Hz, CH ); 8.18, (d, 2H, J = 7.4 Hz, Ar–H); 13.16, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 124.44, 128.71 (2C), 128.95 (4C), 129.83 (2C), 132.26, 133.40, 137.51, 138.88, 142.63, 166.93, 189.32.
4.1.2.2. (Z)-4-(3-Oxo-3-(o-tolyl)prop-1-en-1-yl)benzoic acid (4b)
1H NMR (200 MHz, DMSO-d6): 2.39, (s, 3H, CH3); 7.31–7.38, (m, 2H, Ar–H); 7.44, (d, 1H, JAB = 7.0 Hz, CH ); 7.53, (s, 1H, Ar–H); 7.68, (d, 1H, JBA = 7.0 Hz, CH ); 7.90, (d, 2H, JAB = 8.2 Hz, Ar–H); 7.97, (d, 2H, JBA = 8.2 Hz, Ar–H); 13.13, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 20.13, 125.88, 128.35, 128.60, 128.85 (2C), 129.84 (2C), 131.03, 131.41, 132.32, 136.76, 138.55, 138.62, 143.24, 166.89, 194.77.
4.1.2.3. (E)-4-(3-Oxo-3-(m-tolyl)prop-1-en-1-yl)benzoic acid (4c)
1H NMR (200 MHz, DMSO-d6): 2.42, (s, 3H, CH3); 7.42–7.51, (m, 2H, Ar–H); 7.77, (d, 1H, JAB = 15.6 Hz, CH ); 7.96–8.04, (m, 6H, Ar–H); 8.05, (d, 1H, JBA = 7.0 Hz, CH ); 13.81, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 20.99, 124.50, 125.92, 128.79, 128.95 (2C), 129.11, 129.81 (2C), 132.22, 134.01, 137.55, 138.37, 138.91, 142.45, 166.93, 189.30.
4.1.2.4. (E)-4-(3-Oxo-3-(p-tolyl)prop-1-en-1-yl)benzoic acid (4d)
1H NMR (200 MHz, DMSO-d6): 2.40, (s, 3H, CH3); 7.38, (d, 2H, JAB = 7.8 Hz, Ar–H); 7.78, (d, 1H, JAB = 15.6 Hz, CH ); 7.99, (s, 4H, Ar–H); 8.03, (d, 1H, JBA = 15.6 Hz, CH ); 8.09, (d, 2H, JBA = 7.8 Hz, Ar–H); 13.09, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 21.36, 124.42, 128.92 (2C), 128.98 (2C), 129.55 (2C), 129.88 (2C), 132.21, 135.04, 139.00, 142.32, 143.96, 167.02, 188.70.
4.1.2.5. (Z)-4-(3-(2-Methoxyphenyl)-3-oxoprop-1-en-1-yl)benzoic acid (4e)
1H NMR (200 MHz, DMSO-d6): 3.88, (s, 3H, OCH3); 7.07, (t, 1H, J = 7.2 Hz, Ar–H); 7.20, (d, 1H, JAB = 8.2 Hz, CH ); 7.46–7.63, (m, 3H, Ar–H); 7.53, (d, 1H, JBA = 8.2 Hz, CH ); 7.85, (d, 2H, JAB = 8.2 Hz, Ar–H); 7.97, (d, 2H, JBA = 8.2 Hz, Ar–H); 13.09, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 56.07, 112.60, 120.79, 128.70 (3C), 129.14, 129.85, 130.00 (2C), 132.18, 133.52, 138.94, 141.05, 158.11, 167.00, 192.05.
4.1.2.6. (E)-4-(3-(3-Methoxyphenyl)-3-oxoprop-1-en-1-yl)benzoic acid (4f)
1H NMR (200 MHz, DMSO-d6): 3.85, (s, 3H, OCH3); 7.25, (dd, 1H, J = 8.0 and 2.2 Hz, Ar–H); 7.50, (t, 1H, J = 8.0 Hz, Ar–H); 7.63, (s, 1H, Ar–H); 7.77, (d, 1H, JAB = 15.6 Hz, CH ); 7.79, (d, 1H, J = 8.0 Hz, Ar–H); 7.98, (d, 2H, JAB = 8.6 Hz, Ar–H); 8.02, (d, 2H, JBA = 8.6 Hz, Ar–H); 8.04, (d, 1H, JBA = 15.6 Hz, CH ); 13.09, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 55.56, 113.29, 119.53, 121.33, 124.43, 129.09 (2C), 129.89 (2C), 130.12, 132.31, 138.92, 138.97, 142.78, 159.78, 167.03, 189.04.
4.1.2.7. (E)-4-(3-(4-Methoxyphenyl)-3-oxoprop-1-en-1-yl)benzoic acid (4g)
1H NMR (200 MHz, DMSO-d6): 3.87, (s, 3H, OCH3); 7.09, (d, 2H, J = 9.0 Hz, Ar–H); 7.74, (d, 1H, JAB = 15.6 Hz, CH ); 7.99, (s, 4H, Ar–H); 8.06, (d, 1H, JBA = 15.6 Hz, CH ); 8.19, (d, 2H, JBA = 9.0 Hz, Ar–H); 13.11, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 55.75, 114.25 (2C), 124.43, 128.93 (2C), 129.87 (2C), 130.45, 131.21 (2C), 132.10, 139.12, 141.85, 163.56, 167.03, 187.46.
4.1.2.8. (Z)-4-(3-(2-Fluorophenyl)-3-oxoprop-1-en-1-yl)benzoic acid (4h)
1H NMR (200 MHz, DMSO-d6): 7.36, (d, 1H, JAB = 7.6 Hz, CH ); 7.38–7.44, (m, 1H, Ar–H); 7.52–7.74, (m, 2H, Ar–H); 7.68, (d, 1H, JBA = 7.6 Hz, CH ); 7.81, (t, 1H, J = 7.6 Hz, Ar–H); 7.91, (d, 2H, JAB = 8.2 Hz, Ar–H); 7.99, (d, 2H, JBA = 8.2 Hz, Ar–H); 13.07, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 116.85 (JCF = 22.2 Hz), 125.05 (JCF = 3.3 Hz), 126.79 (JCF = 12.9 Hz), 127.70 (JCF = 3.7 Hz), 128.99 (2C), 129.98 (2C), 130.74, 132.53, 134.67 (JCF = 8.8 Hz), 138.50, 143.20, 160.50 (JCF = 250.6 Hz), 166.94, 188.91.
4.1.2.9. (E)-4-(3-(4-Fluorophenyl)-3-oxoprop-1-en-1-yl)benzoic acid (4i)
1H NMR (200 MHz, DMSO-d6): 7.42, (t, 2H, J = 8.8 Hz, Ar–H); 7.78, (d, 1H, JAB = 15.6 Hz, CH ); 7.96–8.01, (m, 4H, Ar–H); 8.08, (d, 1H, JBA = 15.6 Hz, CH ); 8.28, (m, 2H, Ar–H); 13.15, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 115.99 (2C, JCF = 21.8 Hz), 124.16, 129.08 (2C), 129.87 (2C), 131.80 (2C, JCF = 9.3 Hz), 132.33, 134.21 (JCF = 2.4 Hz), 138.88, 142.83, 165.33 (JCF = 250.8 Hz), 167.00, 187.80.
4.1.2.10. (E)-4-(3-(4-Bromophenyl)-3-oxoprop-1-en-1-yl)benzoic acid (4j)
1H NMR (200 MHz, DMSO-d6): 7.79, (d, 1H, JAB = 15.6 Hz, CH ); 7.80, (d, 2H, JAB = 8.4 Hz, Ar–H); 7.96–8.05, (m, 4H, Ar–H); 8.05, (d, 1H, JBA = 15.6 Hz, CH ); 8.12, (d, 2H, JBA = 8.4 Hz, Ar–H); 13.17, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 124.01, 127.67, 129.10 (2C), 129.84 (2C), 130.77 (2C), 132.02 (2C), 132.45, 136.47, 138.77, 143.15, 166.97, 188.40.
4.1.2.11. (E)-4-(3-(4-Nitrophenyl)-3-oxoprop-1-en-1-yl)benzoic acid (4k)
1H NMR (200 MHz, DMSO-d6): 7.82, (d, 1H, JAB = 15.6 Hz, CH ); 8.00, (s, 4H, Ar–H); 8.06, (d, 1H, JBA = 15.6 Hz, CH ); 8.37, (s, 4H, Ar–H); 13.18, (bs, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 123.97 (3C), 129.22 (2C), 129.85 (2C), 130.09 (2C), 132.64, 138.54, 142.17, 144.09, 150.04, 166.93, 188.39.
4.1.2.12. 4-(3-(Anthracen-1-yl)-3-oxoprop-1-en-1-yl)benzoic acid (4l)
1H NMR (200 MHz, DMSO-d6): 7.54–7.68, (m, 3H, Ar–Hanthracene); 7.74, (d, 1H, JAB = 16.0 Hz, CH ); 7.86, (d, 1H, JBA = 16.0 Hz, CH ); 7.96, (d, 2H, JAB = 8.4 Hz, Ar–H); 8.00, (d, 2H, JBA = 8.4 Hz, Ar–H); 8.10–8.18, (m, 3H, Ar–Hanthracene); 8.35, (d, 1H, J = 8.6 Hz, Ar–Hanthracene); 8.71, (s, 1H, Ar–Hanthracene); 9.13, (s, 1H, Ar–Hanthracene); 13.17, (s, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 124.32, 124.76, 126.43 (2C), 127.25, 127.76, 128.03, 128.54, 128.79, 128.98 (2C), 129.30, 129.92 (2C), 131.31, 131.61, 132.22, 132.35, 133.10, 135.62, 138.80, 143.30, 166.99, 193.52.
4.1.2.13. (E)-4-(3-Oxo-3-(thiophen-2-yl)prop-1-en-1-yl)benzoic acid (4m)
1H NMR (200 MHz, DMSO-d6): 7.33, (t, 1H, J = 4.4 Hz, Ar–Hthiophene); 7.76, (d, 1H, JAB = 15.6 Hz, CH ); 8.00, (s, 4H, Ar–H); 8.01, (d, 1H, JBA = 15.6 Hz, CH ); 8.09, (d, 1H, J = 4.4 Hz, Ar–Hthiophene); 8.38, (d, 1H, J = 4.4 Hz, Ar–Hthiophene); 13.15, (s, 1H, OH); 13C NMR (50 MHz, DMSO-d6): 124.24, 129.06 (3C), 129.88 (2C), 132.31, 134.20, 136.02, 138.77, 141.87, 145.44, 166.99, 181.66.
4.1.2.14. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-oxo-3-phenylprop-1-en-1-yl)benz-amide × H2O (5a)
Beige powder; yield: 0.31 g (68%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 6.87, (d, 1H, J = 8.2 Hz, H-5phenolic); 7.24, (dd, 1H, J = 8.2 and 1.6 Hz, H-6phenolic); 7.39, (d, 1H, J = 1.6 Hz, H-2phenolic); 7.55–7.73, (m, 3H, Ar–H); 7.80, (d, 1H, JAB = 15.6 Hz, CH ); 8.09, (d, 2H, J = 7.4 Hz, Ar–H); 8.12, (d, 1H, JBA = 15.6 Hz, CH ); 8.18–8.22, (m, 4H, Ar–H); 9.45, (s, 1H, OH); 9.61, (s, 1H, OH); 13.13, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 113.86, 116.34, 119.21, 121.58, 124.57, 128.74 (2C), 128.93 (2C), 129.00 (4C), 133.05, 133.41, 137.50, 138.94, 142.48, 146.00, 148.33, 158.58, 162.53, 164.76, 189.26; IR (KBr, cm–1): 3435; 2925; 1666; 1655; 1601; 1533; 1309; 1296; 1219, 747; anal. calcd. for C24H17N3O4S × H2O (461.50 g mol–1): C, 62.46; H, 4.15; N, 9.10; S, 6.95; found: C, 62.54; H, 4.14; N, 9.07; S, 6.97.
4.1.2.15. (Z)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-oxo-3-(o-tolyl)prop-1-en-1-yl)benz-amide × 1.5H2O (5b)
Light yellow powder; yield: 0.41 g (85%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 2.41, (s, 3H, CH3); 6.86, (d, 1H, J = 8.0 Hz, H-5phenolic); 7.24, (d, 1H, J = 8.0 Hz, H-6phenolic); 7.33–7.38, (m, 2H, Ar–H and 1H, H-2phenolic); 7.45, (d, 1H, JAB = 7.0 Hz, CH ); 7.58, (s, 2H, Ar–H); 7.72, (d, 1H, JBA = 7.0 Hz, CH ); 7.98, (d, 2H, JAB = 8.0 Hz, Ar–H); 8.17, (d, 2H, JBA = 8.0 Hz, Ar–H); 9.45, (s, 1H, OH); 9.61, (s, 1H, OH); 13.11, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 20.18, 113.87, 116.34, 119.21, 121.58, 125.87, 128.47, 128.68, 128.88 (2C), 129.04 (2C), 131.06, 131.43, 133.14, 136.83, 138.52, 138.69, 143.04, 146.00, 148.33, 158.60, 162.54, 164.81, 194.68; IR (KBr, cm–1): 3466; 3173; 1667; 1630; 1606; 1532; 1450; 1301; 1275; 807; anal. calcd. for C25H19N3O4S × 1.5H2O (484.53 g mol–1): C, 61.97; H, 4.58; N, 8.67; S, 6.62; found: C, 61.99; H, 4.59; N, 8.65; S, 6.63.
4.1.2.16. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-oxo-3-(m-tolyl)prop-1-en-1-yl)benz-amide (5c)
Yellow powder; yield: 0.38 g (82%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 2.44, (s, 3H, CH3); 6.88, (d, 1H, J = 8.0 Hz, H-5phenolic); 7.26, (dd, 1H, J = 8.0 and 2.0 Hz, H-6phenolic); 7.40, (d, 1H, J = 2.0 Hz, H-2phenolic); 7.44–7.53, (m, 2H, Ar–H); 7.81, (d, 1H, JAB = 15.6 Hz, CH ); 7.96–8.24, (m, 6H, Ar–H); 8.12, (d, 1H, JBA = 15.6 Hz, CH ), 9.46, (s, 1H, OH); 9.62, (s, 1H, OH); 13.14, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 20.98, 113.84, 116.31, 119.17, 121.54, 124.67, 125.94, 128.79, 128.97 (4C), 129.12, 133.01, 134.03, 137.52, 138.37, 138.97, 142.29, 145.97, 148.30, 158.52, 162.50, 164.74, 189.26; IR (KBr, cm–1): 3426; 3169; 1644; 1653; 1607; 1532; 1309; 1297; 1254; 758; anal. calcd. for C25H19N3O4S (457.51 g mol–1): C, 65.63; H, 4.19; N, 9.18; S, 7.01; found: C, 65.61; H, 4.18; N, 9.20; S, 7.02.
4.1.2.17. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-oxo-3-(p-tolyl)prop-1-en-1-yl)benz-amide (5d)
Yellow powder; yield: 0.38 g (84%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 2.71, (s, 3H, CH3); 6.87, (d, 1H, J = 8.2 Hz, H-5phenolic); 7.24, (dd, 1H, J = 8.2 and 1.8 Hz, H-6phenolic); 7.39, (d, 1H, J = 1.8 Hz, H-2phenolic); 7.39, (d, 2H, JAB = 7.8 Hz, Ar–H); 7.78, (d, 1H, JAB = 15.6 Hz, CH ); 8.06–8.13, (m, 4H, Ar–H); 8.11, (d, 1H, JBA = 15.6 Hz, CH ); 8.20, (d, 2H, JBA = 7.8 Hz, Ar–H); 9.46, (s, 1H, OH); 9.61, (s, 1H, OH); 13.13, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 21.31, 113.88, 116.34, 119.20, 121.59, 124.61, 128.96 (6C), 129.49 (2C), 132.97, 135.01, 139.02, 142.09, 143.92, 146.00, 148.33, 158.59, 162.53, 164.78, 188.67; IR (KBr, cm–1): 3434; 3192; 2920; 1668; 1654; 1609; 1533; 1308; 1296; 1210; 759; anal. calcd. for C25H19N3O4S (457.51 g mol–1): C, 65.63; H, 4.19; N, 9.18; S, 7.01; found: C, 65.60; H, 4.20; N, 9.17; S, 7.00.
4.1.2.18. (Z)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-(2-methoxyphenyl)-3-oxoprop-1-en-1-yl)benzamide × 2H2O (5e)
Yellow-green powder; yield: 0.36 g (70%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 3.89, (s, 3H, OCH3); 6.86, (d, 1H, J = 8.2 Hz, H-5phenolic); 7.08, (t, 1H, J = 7.4 Hz, Ar–H); 7.22, (d, 1H, JAB = 8.2 Hz, CH ); 7.23, (d, 1H, J = 8.2 Hz, H-6phenolic); 7.38, (s, 1H, H-2phenolic); 7.54, (d, 1H, JBA = 8.2 Hz, CH ); 7.58, (s, 3H, Ar–H); 7.93, (d, 2H, JAB = 8.2 Hz, Ar–H); 8.17, (d, 2H, JBA = 8.2 Hz, Ar–H); 9.45, (s, 1H, OH); 9.61, (s, 1H, OH); 13.11, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 56.03, 112.56, 113.87, 116.34, 119.22, 120.72, 121.60, 128.62 (2C), 128.73, 129.10 (2C), 129.28, 129.76, 132.99, 133.41, 138.95, 140.84, 146.00, 148.33, 158.06, 158.59, 162.51, 164.81, 192.00; IR (KBr, cm–1): 3434; 3236; 2934; 1661; 1607; 1536; 1448; 1315; 1295; 1245; 747; anal. calcd. for C25H19N3O5S × 2H2O (509.54 g mol–1): C, 58.93; H, 4.55; N, 8.25; S, 6.29; found: C, 58.95; H, 4.57; N, 8.26; S, 6.27.
4.1.2.19. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-(3-methoxyphenyl)-3-oxoprop-1-en-1-yl)benzamide × H2O (5f)
Dark yellow powder; yield: 0.38 g (78%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 3.86, (s, 3H, OCH3); 6.87, (d, 1H, J = 8.0 Hz, H-5phenolic); 7.23–7.27, (m, 1H, Ar–H and 1H, H-6phenolic); 7.39, (s, 1H, H-2phenolic); 7.51, (t, 1H, J = 8.0 Hz, Ar–H); 7.65, (s, 1H, Ar–H); 7.80, (d, 1H, JAB = 15.6 Hz, CH ); 7.82, (d, 1H, J = 8.0 Hz, Ar–H); 8.09, (d, 2H, JAB = 8.6 Hz, Ar–H); 8.10, (d, 1H, JBA = 15.6 Hz, CH ); 8.20, (d, 2H, JBA = 8.6 Hz, Ar–H); 9.45, (s, 1H, OH); 9.61, (s, 1H, OH); 13.13, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 55.54, 113.32, 113.88, 116.35, 119.23, 119.46, 121.30, 121.61, 124.59, 129.04 (4C), 130.06, 133.06, 138.94 (2C), 142.56, 146.01, 148.34, 158.63, 159.74, 162.55, 164.78, 189.00; IR (KBr, cm–1): 3434; 2937; 1663; 1607; 1538; 1448; 1317; 1293; 1259; 759; anal. calcd. for C25H19N3O5S × H2O (491.52 g mol–1): C, 61.09; H, 4.31; N, 8.55; S, 6.52; found: C, 61.11; H, 4.29; N, 8.57; S, 6.53.
4.1.2.20. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-(4-methoxyphenyl)-3-oxoprop-1-en-1-yl)benzamide × 0.5H2O (5g)
Light yellow powder; yield: 0.34 g (70%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 3.88, (s, 3H, OCH3); 6.87, (d, 1H, J = 8.2 Hz, H-5phenolic); 7.10, (d, 2H, JAB = 9.0 Hz Ar–H); 7.24, (dd, 1H, J = 8.2 and 2.0 Hz, H-6phenolic); 7.39, (d, 1H, J = 2.0 Hz, H-2phenolic); 7.76, (d, 1H, JAB = 15.6 Hz, CH ); 8.08, (d, 2H, JAB = 8.4 Hz, Ar–H); 8.13, (d, 1H, JBA = 15.6 Hz, CH ); 8.20, (d, 2H, JBA = 8.4 Hz, Ar–H); 8.21, (d, 2H, JBA = 9.0 Hz, Ar–H); 9.46, (s, 1H, OH); 9.61, (s, 1H, OH); 13.12, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 55.71, 113.84, 114.20 (2C), 116.32, 119.18, 121.56, 124.62, 128.46, 128.89 (2C), 128.98 (2C), 130.42, 131.17 (2C), 139.14, 141.61, 145.98, 148.31, 158.48, 162.52, 163.53, 164.78, 187.40; IR (KBr, cm–1): 3406; 3173; 2940; 1670; 1654; 1605; 1593; 1533; 1307; 1263; 1172; 765; anal. calcd. for C25H19N3O5S × 0.5H2O (482.52 g mol–1): C, 62.23; H, 4.18; N, 8.71; S, 6.64; found: C, 62.25; H, 4.19; N, 8.72; S, 6.65.
4.1.2.21. (Z)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-(2-fluorophenyl)-3-oxoprop-1-en-1-yl)benzamide × H2O (5h)
Yellow powder; yield: 0.26 g (54%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 6.86, (d, 1H, J = 8.2 Hz, H-5phenolic); 7.24, (dd, 1H, J = 8.2 and 1.6 Hz, H-6phenolic); 7.37, (d, 1H, JAB = 7.6 Hz, CH ); 7.39–7.44, (m, 1H, Ar–H and 1H, H-2phenolic); 7.58–7.80 (m, 2H, Ar–H and 1H, CH ), 7.83 (t, 1H, J = 7.6 Hz, Ar–H); 7.99, (d, 2H, JAB = 8.2 Hz, Ar–H); 8.18, (d, 1H, JBA = 8.2 Hz, Ar–H); 9.46, (s, 1H, OH); 9.61, (s, 1H, OH); 13.14, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 113.86, 116.33, 116.78 (JCF = 22.2 Hz), 119.21, 121.57, 124.97 (JCF = 3.3 Hz), 126.76 (JCF = 12.8 Hz), 127.81 (JCF = 3.7 Hz), 128.92 (2C), 129.10 (2C), 130.70 (JCF = 2.1 Hz), 133.34, 134.59 (JCF = 8.8 Hz), 138.51, 142.97, 146.00, 148.33, 158.64, 160.46 (JCF = 250.8 Hz), 162.51, 164.79, 188.84; IR (KBr, cm–1): 3405; 2928; 1671; 1662; 1611; 1539; 1451; 1317; 1293; 1268; 755; anal. calcd. for C24H16N3O4SF × H2O (479.49 g mol–1): C, 60.11; H, 3.96; N, 8.76; S, 6.69; found: C, 60.13; H, 3.95; N, 8.78; S, 6.70.
4.1.2.22. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-(4-fluorophenyl)-3-oxoprop-1-en-1-yl)benzamide × 2H2O (5i)
Yellow powder; yield: 0.31 g (63%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 6.87, (d, 1H, J = 8.2 Hz, H-5phenolic); 7.24, (dd, 1H, J = 8.2 and 1.6 Hz, H-6phenolic); 7.38, (d, 1H, J = 1.6 Hz, H-2phenolic); 7.42, (t, 2H, J = 8.8 Hz, Ar–H); 7.81, (d, 1H, JAB = 15.6 Hz, CH ); 8.09, (d, 2H, JAB = 8.4 Hz, Ar–H); 8.14, (d, 1H, JBA =15.6 Hz, CH ); 8.20, (d, 2H, JBA = 8.4 Hz, Ar–H); 8.31, (m, 2H, Ar–H); 9.45, (s, 1H, OH); 9.61, (s, 1H, OH); 13.13, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 113.84, 115.93 (2C, JCF = 21.6 Hz), 116.31, 119.18, 121.55, 124.32, 128.99 (2C), 129.02 (2C), 131.76 (2C, JCF = 9.4 Hz), 133.06, 134.17 (JCF = 2.6 Hz), 138.88, 142.61, 145.98, 148.31, 158.50 (JCF = 6.7 Hz), 162.51, 164.73, 165.28 (JCF = 250.8 Hz), 187.75; IR (KBr, cm–1): 3225; 3071; 2951; 1668; 1654; 1603; 1538; 1508; 1308; 1220; 831; anal. calcd. for C24H16N3O4SF × 2H2O (497.50 g mol–1): C, 57.94; H, 4.05; N, 8.45; S, 6.44; found: C, 57.92; H, 4.06; N, 8.43; S, 6.45.
4.1.2.23. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-(4-bromophenyl)-3-oxoprop-1-en-1-yl)benzamide × 2H2O (5j)
Yellow-green powder; yield: 0.49 g (87%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 6.87, (d, 1H, J = 7.6 Hz, H-5phenolic); 7.24, (d, 1H, J = 7.6 Hz, H-6phenolic); 7.39, (s, 1H, H-2phenolic); 7.79, (d, 2H, J = 8.4 Hz, Ar–H); 7.80, (d, 1H, JAB = 15.6 Hz, CH ); 8.05–8.21, (m, 6H, Ar–H and 1H, CH ); 9.46, (s, 1H, OH); 9.61, (s, 1H, OH); 13.13, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 113.85, 116.32, 119.19, 121.55, 124.20, 127.63, 129.00 (2C), 129.08 (2C), 130.75 (2C), 131.99 (2C), 133.14, 136.46, 138.83, 142.95, 145.99, 148.32, 158.59, 162.53, 164.69, 188.37; IR (KBr, cm–1): 3406; 3033; 2923; 1662; 1655; 1600; 1584; 1534; 1305; 1214; 759; anal. calcd. for C24H16N3O4SBr × 2H2O (558.41 g mol–1): C, 51.62; H, 3.61; N, 7.52; S, 5.74; found: C, 51.64; H, 3.60; N, 7.54; S, 5.75.
4.1.2.24. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-(4-nitrophenyl)-3-oxoprop-1-en-1-yl)benzamide × 4H2O (5k)
Brown powder; yield: 0.30 g (53%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 6.86, (d, 1H, J = 6.4 Hz, H-5phenolic); 7.24, (d, 1H, J = 6.4 Hz, H-6phenolic); 7.38, (s, 1H, H-2phenolic); 7.86, (d, 1H, JAB = 15.6 Hz, CH ); 8.09–8.19, (m, 4H, Ar–H and 1H, CH ); 8.40, (s, 4H, Ar–H); 9.45, (s, 1H, OH); 9.61, (s, 1H, OH); 13.16, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 113.84, 116.31, 119.18, 121.54, 123.95 (2C), 124.22, 129.02 (2C), 129.23 (2C), 130.09 (2C), 133.36, 138.61, 142.18, 143.91, 145.98, 148.33, 150.07, 158.59, 162.51, 164.71, 188.42; IR (KBr, cm–1): 3404; 3165; 2930; 1664; 1655; 1607; 1590; 1523; 1306; 1298; 1212; 747; anal. calcd. or C24H16N4O6S × 4H2O (560.54 g mol–1): C, 51.43; H, 4.32; N, 10.00; S, 5.72; found: C, 51.45; H, 4.31; N, 9.98; S, 5.73.
4.1.2.25. (E)-4-(3-(Anthracen-1-yl)-3-oxoprop-1-en-1-yl)-N-(5-(3,4-dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)benzamide × H2O (5l)
Dark orange powder; yield: 0.43 g (76%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 6.87, (d, 1H, J = 8.2 Hz, H-5phenolic); 7.25, (d, 1H, J = 8.2 Hz, H-6phenolic); 7.39, (s, 1H, H-2phenolic); 7.55–7.70, (m, 3H, Ar–Hanthracene), 7.78, (d, 1H, JAB = 15.8 Hz, CH ); 7.92, (d, 1H, JBA = 15.8 Hz, CH ); 7.98–8.22, (m, 4H, Ar–H and 3H, Ar–Hanthracene); 8.37, (d, 1H, J = 8.4 Hz, Ar–Hanthracene); 8.73, (s, 1H, Ar–Hanthracene); 9.16, (s, 1H, Ar–Hanthracene); 9.46, (s, 1H, OH); 9.62, (s, 1H, OH); 13.14, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 113.90, 116.36, 119.24, 121.62, 124.27, 124.72, 126.38 (2C), 127.21, 127.72, 127.99, 128.65, 128.74 (2C), 128.93 (2C), 129.05, 131.28, 131.58, 132.19, 133.10, 135.57, 138.82, 143.06, 146.02, 148.35, 158.63, 162.54, 164.80, 193.43; IR (KBr, cm–1): 3405; 3047; 2930; 1655; 1607; 1538; 1510; 1302; 1290; 1255; 1201; 749; anal. calcd for C32H21N3O4S × H2O (561.62 g mol–1): C, 68.43; H, 4.13; N, 7.48; S, 5.71; found: C, 68.40; H, 4.14; N, 7.50; S, 5.73.
4.1.2.26. (E)-N-(5-(3,4-Dihydroxyphenyl)-1,3,4-thiadiazol-2-yl)-4-(3-oxo-3-(thiophen-2-yl)prop-1-en-1-yl)benzamide (5m)
Yellow powder; yield: 0.28 g (62%); mp: >250 °C; 1H NMR (200 MHz, DMSO-d6): 6.87, (d, 1H, J = 8.2 Hz, H-5phenolic); 7.24, (dd, 1H, J = 8.2 and 1.8 Hz, H-6phenolic); 7.34, (t, 1H, J = 4.4 Hz, Ar–Hthiophene); 7.39, (d, 1H, J = 1.8 Hz, H-2phenolic); 7.79, (d, 1H, JAB = 15.6 Hz, CH ); 8.06, (d, 1H, JBA = 15.6 Hz, CH ); 8.08, (d, 2H, JAB = 8.4 Hz, Ar–H); 8.10, (d, 1H, J = 4.4 Hz, Ar–Hthiophene); 8.20, (d, 2H, JBA = 8.4 Hz, Ar–H); 8.41, (d, 1H, J = 4.4 Hz, Ar–Hthiophene); 9.47, (s, 1H, OH); 9.62, (s, 1H, OH); 13.13, (s, 1H, NH); 13C NMR (50 MHz, DMSO-d6): 113.85, 116.33, 119.20, 121.55, 124.44, 129.01 (5C), 133.07, 134.18, 135.99, 138.80, 141.66, 145.39, 145.99, 148.32, 158.55, 162.53, 164.81, 181.63; IR (KBr, cm–1): 3406; 2927; 1671; 1654; 1595; 1540; 1413; 1317; 1292; 1189; 760; anal. calcd. for C22H15N3O4S2 (449.51 g mol–1): C, 58.78; H, 3.36; N, 9.35; S, 14.26; found: C, 58.80; H, 3.34; N, 9.36; S, 14.21.
4.2. Biology
4.2.1. Cytotoxic activity
Cytotoxic activity of newly synthesized 1,3,4-thiadiazole–chalcone hybrids containing the antioxidant phenolic moiety was evaluated using three human malignant cell lines: cervical adenocarcinoma HeLa, acute promyelocytic leukemia HL-60, lung carcinoma A549, and human normal lung fibroblasts MRC-5, as described in our previous studies.20 All tested cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). HeLa (2000 cells per well), A549 (5000 cells per well), and MRC-5 cells (5000 cells per well) were seeded into 96-well microtiter plates and after 20 h five different concentrations of the compounds (ranging from 6.25 μM to 100 μM or 3.125 μM to 50 μM) were added to the cells. The control cell samples were incubated in nutrient medium only. HL-60 cells (7000 cells per well), were seeded 2 h before the addition of solutions of the compounds. Stock solutions of the compounds were made in DMSO at a concentration of 5 mM. Cell survival was measured by MTT assay after treatment that lasted 72 h, according to the standard procedure first described by Mosmann,52 and which was modified by Ohno and Abe,53 as described previously in more detail.20 Concentrations of the compounds dissolved in DMSO and further diluted in nutrient medium were carefully chosen in order to eliminate any toxic side effect of DMSO on all tested human cell lines. The maximum final concentrations of DMSO did not exceed 1%.
The chemotherapy drug cisplatin was used as a positive control. Each of the three independent experiments was performed in triplicate.
4.2.2. DPPH free radical scavenging assay
Determination of DPPH free radical scavenging activity of the studied compounds was conducted according to a method described by Kontogiorgis et al.54 Briefly, 1 mL (0.05 mM) of DPPH solution in methanol was mixed with an equivalent volume of the tested compound (20 μL of compound solution in DMSO and 980 μL of methanol). The sample was stored in the dark at room temperature. Absorbance was acquired at 517 nm after an incubation period of 30 min. Methanol was taken as control. IC50 values represent the concentration necessary to obtain 50% of a maximum scavenging activity. Ascorbic acid was used as positive control. The results are presented as mean ± SD calculated from independent triplicate experiments using Microsoft Excel software.
4.2.3. Determination of intracellular ROS levels
Human HeLa cells were incubated with subtoxic IC20 concentrations of selected compounds 5a, 5c, 5f, and 5m (applied concentration was 10 μM for each compound) for 24 h. Afterwards, the HeLa cells were collected, washed with phosphate buffered saline (PBS) and incubated in a solution of 30 μM 2′,7′-dichlorodihydrofluorescein diacetate (Sigma-Aldrich) in PBS for 45 min at 37 °C, according to a standard experimental procedure which we described previously.55 The cells were then washed with PBS. Part of the treated cell samples was analysed immediately, while the other part of treated cells samples was exposed to 10 mM hydrogen peroxide solution (H2O2) for 30 min at 37 °C. After 30 min, these cells were washed with PBS and collected. The intensity of green fluorescence emitted by the dichlorofluorescein was determined on a FACSCalibur flow cytometer (BD Biosciences Franklin Lakes, NJ, USA). The data (20 000 events acquired for each cell sample) were analysed using CELLQuest software (BD Biosciences).
4.2.4. Cell cycle analysis by flow cytometry
Human cervical carcinoma HeLa cells were incubated with compounds 5a, 5c, 5f, and 5m for 24 h (applied concentrations corresponded to IC50 and 2IC50 concentrations which were determined after 72 h treatment). Following 24 h incubation, the cells were collected, washed with PBS, and fixed in 70% ethanol, according to standard protocol.56 Cell samples were stored at –20 °C for at least one week before staining. Afterwards, HeLa cells were collected by centrifugation, washed and resuspended in PBS containing RNase A, and incubated for 30 min at 37 °C. Subsequently propidium iodide staining solution was added to the cells. Percentages of HeLa cells within specific phases of the cell cycle were assessed using a BD FACSCalibur flow cytometer. Analyses of acquired data (10 000 events collected for each gated cell sample) were performed using CELLQuest software. Cell cycle distribution data are presented as mean ± S.D. of three independent experiments. Statistical significance of differences between the control and cell samples exposed to tested compounds were evaluated using one-way ANOVA with Dunnett's post-test and p values below 0.05 were considered statistically significant.
4.2.5. Identification of target caspases
To investigate whether the examined compounds could induce apoptosis in treated HeLa cells the percentages of subG1 cells in samples pretreated with specific caspase inhibitors were determined, as described in our previous research.20 The HeLa cells were pretreated for 2 h with 40 μM concentrations of specific peptide caspase inhibitors: Z-DEVD-FMK, a caspase-3 inhibitor, Z-IETD-FMK, a caspase-8 inhibitor, and Z-LEHD-FMK, a caspase-9 inhibitor (R&D Systems, Minneapolis, USA).
4.2.6. Gene and microRNA expression analyses
HeLa cells were seeded into 75 cm2 cell culture flasks (4 × 106 cells/flask). After 2 h, the cells were treated with low subtoxic IC20 concentrations of compounds 5a, 5c, 5f, and 5m for 24 h (10 μM for each compound). Control cells were grown in nutrient medium only. After 24 h incubation, the cells were collected, washed, and the cell samples were stored at –80 °C until further analyses. Total RNA was extracted with TriReagent (Sigma), according to manufacturer's protocol. Gene expression was quantified by a two-step reverse transcription reaction followed by real-time quantitative PCR (RT-qPCR) with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems by Thermo Fisher Scientific, Vilnius, Lithuania). Reaction of cDNA amplification was performed with MMP2 (Hs01548727_m1), MMP9 (Hs00957562_m1), VEGFA (Hs00900055_m1), and TIMP3 (Hs00165949_m1) assays, using TaqMan technology.
Stem-loop reverse transcription and RT-qPCR were used for miR-21 (ID 000397), miR-133b (ID 002247), miR-155 (ID 002623), and miR-206 (ID 000510) expression analysis.
Gene and microRNA expression values were normalized to GAPDH (Hs02758991_g1), and small nuclear RNA-RNU6 B (ID001093) and calculated by comparative ΔΔCt method, with 7500 System SDS software (Applied Biosystems, Foster City, California, USA).
4.2.7. Endothelial cell tube formation assay
Antiangiogenic effects of the compounds were explored using human umbilical vein endothelial EA.hy926 cells.57,58 The EA.hy926 cells seeded on Corning® Matrigel® basement membrane matrix (Corning: cat. number 356234) were exposed to subtoxic IC20 concentrations of the four tested compounds (compounds 5a, 5c, 5f (6.5 μM for each compound), and 5m (8 μM) for 20 h. After treatment that lasted 20 h, photomicrographs of EA.hy926 cells were captured under an inverted phase-contrast microscope.
4.2.8. Interaction with DNA
4.2.8.1. Absorption spectral measurements
Calf thymus DNA (lyophilized, highly polymerized, obtained from Serva, Heidelberg) (CT-DNA) was used for these experiments. A stock solution was prepared by dissolving the DNA in Tris buffer (10 mM Tris-HCl pH 7.9) overnight at 4 °C. This stock solution was stored at 4 °C and was stable for several days. A solution of CT-DNA in water gave a ratio of UV absorbance at 260 and 280 nm, A260/A280 of 1.89–2.01, indicating that DNA was sufficiently free of protein. The concentration of DNA (3.16 mg mL–1) was determined from UV absorbance at 260 nm using the extinction coefficient ε260 = 6600 M–1 cm–1.59 1,3,4-Thiadiazole-chalcone hybrids 5a, 5c, 5f, and 5m were dissolved in dimethyl sulfoxide in concentrations of 2 mM. These solutions were used as stock solutions.
For UV-vis measurements, a small volume of a stock solution of a compound (20 μL) was added to DNA solution (10 μL of CT-DNA) and the volume was adjusted up to 1 mL with 40 mM bicarbonate buffer, pH 8.4. Reaction mixtures were incubated at 37 °C for 90 min with occasional vortexing. UV-vis spectra were recorded on a UV-1800 Shimadzu UV/visible spectrophotometer operating from 200 to 800 nm in 1.0 cm quartz cells. Spectra of the compounds of the same concentrations were also recorded, as well as spectra of CT-DNA.
The percentage of hyperchromism or hypocromism was determined from eqn (1):
| {[(εDNA + εCOMP) – εB]/(εDNA + εCOMP)} × 100 | 1 |
where εDNA is the extinction coefficient of CT-DNA, εCOMP is the extinction coefficient of free compound, and εB is the extinction coefficient of the bound compound complex.
The absorbance titrations were performed at a fixed concentration of the compound with gradually increasing concentration of double stranded CT-DNA. Absorbance at 259 nm was monitored for each concentration of DNA. The binding constant Kb was determined using eqn (2):60
| [DNA] × (εa – εf)–1 = [DNA] × (εb – εf)–1 + Kb–1 × (εb – εf)–1, | 2 |
where εa, εf, εb are absorbance/[compound], extinction coefficient of the free compound, and the extinction coefficient of the bound compound, respectively. A plot of [DNA]/(εa – εf) versus [DNA] gave a slope and an intercept equal to 1/(εa – εf) and (1/Kb)(1/(εb – εf)), respectively. The binding constant Kb is calculated from the ratio of the slope to the intercept.
4.2.8.2. Fluorescence measurements
Competitive interactions of 1,3,4-thiadiazole–chalcone hybrids 5a, 5c, 5f, and 5m and the fluorescence probe Hoechst 33258 (H), with CT-DNA were studied by measuring the change of fluorescence intensity of H–DNA solution after addition of one of the compounds. Reaction mixtures containing 100 μM of CT-DNA (calculated per phosphate) in 1 mL of 40 mM bicarbonate solution (pH 8.4) were pretreated with 1.5 μL of 1% H probe solution (28 μM final concentration) (in separate experiments) for 20 min and the mixture was analyzed by fluorescence. Then, increasing concentrations of the compounds were successively added and changes in the fluorescence intensity were measured using a Thermo Scientific Lumina Fluorescence spectrometer (Finland) equipped with a 150 W xenon lamp. The slits on the excitation and emission beams were fixed at 10 nm. All measurements were performed by excitation at 350 nm in the range of 390–550 nm. The control was H–CT-DNA solution. Compounds 5a, 5c, 5f, and 5m did not have fluorescence under applied conditions. The obtained fluorescence quenching data were analyzed according to the Stern–Volmer eqn (3):61
| I0/I = 1 + Kr | 3 |
where I0 and I represent the fluorescence intensities of H–CT-DNA in absence and presence of the compounds, respectively, K is quenching constant, and r is ratio of the bound concentration of a probe to the bound concentration of DNA (r = [compound]/[CT-DNA]. The K value is calculated from the ratio of the slope to the intercept from the plot of I0/I versusr.
Primary spectra of all spectrometric measurements were imported into OriginPro 9.0 and were processed by this software package.
4.2.8.3. Experiments with plasmid DNA
Plasmid pUC19 (2686 bp in length, purchased from Sigma-Aldrich, USA) was prepared by its transformation in chemically competent cells Escherichia coli (E. coli) strain XL1 blue. Amplification of the clone was done according to the protocol for growing E. coli culture overnight in LB medium at 37 °C (ref. 62) and purification was performed using a Qiagen Plasmid plus Maxi kit. Finally, DNA was eluted in 10 mM Tris-HCl buffer and stored at –20 °C. The concentration of plasmid DNA (0.460 μg μL–1) was determined by measuring the absorbance of the DNA-containing solution at 260 nm. One optical unit corresponds to 50 μg mL–1 of double stranded DNA.
Plasmid DNA (1 μL, 460 ng μL–1) was incubated with increasing volumes of 2 mM stock solution in DMSO of each compound (0.5, 1, 1.5, 2, 2.5, 3, and 4 μL) in a 20 μL reaction mixture in 40 mM bicarbonate buffer (pH 8.4) at 37 °C, for 90 minutes. DMSO had no effect on DNA conformation under applied concentrations. The reaction mixtures were vortexed from time to time. The reaction was terminated by short centrifugation at 10 000 rpm and addition of 5 μL of loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol FF and 30% glycerol in TAE buffer, pH 8.24 (40 mM Tris-acetate, 1 mM EDTA)), and analyzed by agarose electrophoresis.
Interactions of the tested compounds with pUC19 in presence of Fe(ii) and Fe(iii) were done as follows: solutions of 20 μL volume consisting of 1 μL or 2 μL FeSO4 (8 mM, freshly prepared in a sterile water) or 1 μL or 2 μL FeCl3 (8 mM, freshly prepared in a sterile water) with 4 μL of 2 mM solution of 1,3,4-thiadiazole–chalcone hybrids 5a, 5c, 5f, and 5m in bicarbonate buffer were incubated at 37 °C, for 60 minutes, then 1 μL of pUC19 was added to the reaction mixtures and incubated further for 90 minutes. The reaction mixtures were vortexed from time to time and terminated by short centrifugation at 10 000 rpm and addition of 5 μL of loading buffer and analyzed by agarose electrophoresis.
4.2.8.4. Agarose electrophoresis
Samples were subjected to electrophoresis on 1% agarose gel (Amersham Pharmacia-Biotech, Inc) prepared in TAE buffer pH 8.24. The electrophoresis was performed at a constant voltage (80 V) until bromophenol blue had passed through 75% of the gel. A Submarine Mini-gel Electrophoresis Unit (Hoeffer HE 33) with an EPS 300 power supply was used. After electrophoresis, the gel was stained for 30 min by soaking it in an aqueous ethidium bromide solution (0.5 μg mL–1). The stained gel was illuminated under a UV transilluminator (Vilber-Lourmat, France) at 312 nm and photographed with a Nikon Coolpix P340 Digital Camera through filter DEEP YELLOW 15 (TIFFEN, USA).
4.2.8.5. Comet assay
Human normal MRC-5 cells were treated with four concentrations of selected compounds 5a, 5c, 5f, and 5m (applied concentrations were 25 μM, 12.5 μM, 6.25 μM, and 3.125 μM for each compound) for 24 h. The IC20 subtoxic concentration was 25 μM for all four compounds, as determined by a MTT test after 24 h treatment. After incubation, the cells were collected, washed with phosphate buffered saline (PBS), suspended in freezing medium (RPMI with 10% DMSO and 20% FCS) and the cell samples were frozen at –80 °C.
The medium-throughput version of the single-cell gel electrophoresis assay was used to evaluate DNA damage as previously described63 with some modifications. Frozen MRC-5 cells were thawed by addition of 1 mL of PBS to a frozen 0.5 mL aliquot and as soon as the sample was thawed the suspended cells were centrifuged for 10 min at 2000 rpm at 4 °C. The pellet was suspended in PBS and the washing step was repeated. The number of cells was adjusted to 2.5 × 105 mL–1 with PBS and 30 μL of cell suspension was mixed with 140 μL of 1% LMP agarose at 37 °C. Twelve drops (10 μL) of agarose-cell suspension was placed on a NMP agarose coated slide using a template provided on the metal base of the 12 gel chamber. Cells were lysed by placing the slides in 2.5 M NaCl, 0.1 M Na2EDTA, 10 mM Tris with 1% Triton X-100 pH 10 for 1 h at 4 °C. Then, slides were placed in a horizontal gel electrophoresis tank in electrophoresis solution (0.3 M NaOH, 0.001 M Na2EDTA) for 20 min. Electrophoresis was carried out for 30 min at a voltage gradient of 1 V cm–1 across the platform at 4 °C. Neutralization of the slides was performed by washing them for 10 min in PBS in a staining jar at 4 °C. Slides were then fixed by placing them in 70% ethanol for 10 min followed by 10 min incubation in absolute ethanol. DNA was stained by immersing the slides in a staining jar with SYBR Gold (Invitrogen) for 30 min in the dark. Staining solution was prepared according to the manufacturer's instructions in 10 mM Tris, 1 mM EDTA buffer, pH 8. Stained slides were rinsed twice with water and left to dry in the dark. Scoring of the comets was carried out using a semi-automated image analysis system (Comet Assay IV; Perceptive Instruments). On each gel 50 nucleoids were analyzed, and the results were expressed as percentage of tail intensity (% of DNA in tail). All experiments were performed in triplicate. PBS was used as a negative control. The median% tail DNA for 50 comets was calculated for each of the duplicate gels in the experiments; the mean of the two median values was then calculated. The mean percentage of DNA in the tail was calculated from independent triplicate experiments using Microsoft Excel software.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors are grateful to the Ministry of Science and Technological Development of the Republic of Serbia for financial support (Grant No. 172016, 172055 and 175011).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8md00316e
References
- Abbot V., Sharma P., Dhiman S., Noolvi M. N., Patel H. M., Bhardwaj V. RSC Adv. 2017;7:28313–28349. [Google Scholar]
- Mahapatra D. K., Bharti S. K., Asati V. Eur. J. Med. Chem. 2015;98:69–114. doi: 10.1016/j.ejmech.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Kerru N., Singh P., Koorbanally N., Raj R., Kumar V. Eur. J. Med. Chem. 2017;142:179–212. doi: 10.1016/j.ejmech.2017.07.033. [DOI] [PubMed] [Google Scholar]
- Wei H., Ruan J., Zhang X. RSC Adv. 2016;6:10846–10860. [Google Scholar]
- Pingaew R., Saekee A., Mandi P., Nantasenamat C., Prachayasittikul S., Ruchirawat S., Prachayasittikul V. Eur. J. Med. Chem. 2014;85:65–76. doi: 10.1016/j.ejmech.2014.07.087. [DOI] [PubMed] [Google Scholar]
- Yadav P., Lal K., Kumar A., Guru S. K., Jaglan S., Bhushan S. Eur. J. Med. Chem. 2017;126:944–953. doi: 10.1016/j.ejmech.2016.11.030. [DOI] [PubMed] [Google Scholar]
- Mizuno C. S., Paul S., Suh N., Rimando A. M. Bioorg. Med. Chem. Lett. 2010;20:7385–7387. doi: 10.1016/j.bmcl.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim G. A. M., Guimarães T. T., do Carmo M., Pinto F. R., Cavalcanti B. C., de Farias K. M., Pessoa C., Gatto C. C., Nair D. K., Namboothiri I. N. N., da Silva Júnior E. N. Med. Chem. Commun. 2015;6:120–130. [Google Scholar]
- Shankaraiah N., Siraj K. P., Nekkanti S., Srinivasulu V., Sharma P., Senwar K. R., Sathish M., Vishnuvardhan M. V. P. S., Ramakrishna S., Jadala C., Nagesh N., Kamal A. Bioorg. Chem. 2015;59:130–139. doi: 10.1016/j.bioorg.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Xie L., Zhai X., Ren L., Meng H., Liu C., Zhu W., Zhao Y. Chem. Pharm. Bull. 2011;59:984–990. doi: 10.1248/cpb.59.984. [DOI] [PubMed] [Google Scholar]
- Abdel-Aziz M., Park S.-E., El-Din G., Abuo-Rahma A. A., Sayed M. A., Kwon Y. Eur. J. Med. Chem. 2013;69:427–438. doi: 10.1016/j.ejmech.2013.08.040. [DOI] [PubMed] [Google Scholar]
- Marković V., Debeljak N., Stanojković T., KolundŽija B., Sladić D., Vujčić M., Janović B., Tanić N., Perović M., Tešić V., Antić J., Joksović M. D. Eur. J. Med. Chem. 2015;89:401–410. doi: 10.1016/j.ejmech.2014.10.055. [DOI] [PubMed] [Google Scholar]
- Sribalan R., Banuppriya G., Kirubavathi M., Jayachitra A., Padmini V. Bioorg. Med. Chem. Lett. 2016;26:5624–5630. doi: 10.1016/j.bmcl.2016.10.075. [DOI] [PubMed] [Google Scholar]
- Shi H.-B., Zhang S.-J., Ge Q.-F., Guo D.-W., Cai C.-M., Hu W.-X. Bioorg. Med. Chem. Lett. 2010;20:6555–6559. doi: 10.1016/j.bmcl.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Wan M., Xu L., Hua L., Li A., Li S., Lu W., Pang Y., Cao C., Liu X., Jiao P. Bioorg. Chem. 2014;54:38–43. doi: 10.1016/j.bioorg.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Hu Y., Li C.-Y., Wang X.-M., Yang Y.-H., Zhu H.-L. Chem. Rev. 2014;114:5572–5610. doi: 10.1021/cr400131u. [DOI] [PubMed] [Google Scholar]
- Jain A. K., Sharma S., Vaidya A., Ravichandran V., Agrawal R. K. Chem. Biol. Drug Des. 2013;81:557–576. doi: 10.1111/cbdd.12125. [DOI] [PubMed] [Google Scholar]
- Haider S., Alam M. S., Hamid H. Eur. J. Med. Chem. 2015;92:156–177. doi: 10.1016/j.ejmech.2014.12.035. [DOI] [PubMed] [Google Scholar]
- Halliwell B. and Gutteridge J. M. in Free Radicals in Biology and Medicine, Oxford University Press, Midsomer Norton, Avon, England, 3rd edn, 1999. [Google Scholar]
- Jakovljević K., Matić I. Z., Stanojković T., Krivokuća A., Marković V., Joksović M. D., Mihailović N., Nićiforović M., Joksović Lj. Bioorg. Med. Chem. Lett. 2017;27:3709–3715. doi: 10.1016/j.bmcl.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Ullrich T., Ghobrial M., Weigand K., Marzinzik A. L. Synth. Commun. 2007;37:1109–1119. [Google Scholar]
- Jovanović B. ž., Mišić-Vuković M., Marinković A. D., Csanádi J. J. Mol. Struct. 1999;482–483:371–374. [Google Scholar]
- Tanaka T., Kojima T., Kawamori T., Mori H. Cancer. 1995;75:1433–1439. doi: 10.1002/1097-0142(19950315)75:6+<1433::aid-cncr2820751507>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Markaryan Sh. A., Tavadyan L. A., Kocharyan G. G., Shahinyan G. A. Russ. Chem. Bull., Int. Ed. 2013;62:1625–1629. [Google Scholar]
- Valgimigli L., Banks J. T., Ingold K. U., Lusztyk J. J. Am. Chem. Soc. 1995;117:9966–9971. [Google Scholar]
- Procházková A., Boušová I., Wilhelmová N. Fitoterapia. 2011;82:513–523. doi: 10.1016/j.fitote.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Glorieux C., Dejeans N., Sid B., Beck R., Buc Calderon P., Verrax J. Biochem. Pharmacol. 2011;82:1384–1390. doi: 10.1016/j.bcp.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Das M., Manna K. J. Toxicol. 2016;2016:7651047. doi: 10.1155/2016/7651047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pan X., Cobb G. P., Anderson T. A. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Song B., Wang C., Liu J., Wang X., Lv L., Wei L., Xie L., Zheng Y., Song X. J. Exp. Clin. Cancer Res. 2010;29:29. doi: 10.1186/1756-9966-29-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partyka R., Gonciarz M., Jałowiecki P., Kokocińska D., Byrczek T. Med. Sci. Monit. 2012;18:BR130–BR134. doi: 10.12659/MSM.882614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashaei E., Pashaei E., Ahmady M., Ozen M., Aydin N. PLoS One. 2017;12:e0179543. doi: 10.1371/journal.pone.0179543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. H., Qin Y. E., Tang W. F., Tao J., Song H., Zuo M. Cancer Cell Int. 2017;17:63. doi: 10.1186/s12935-017-0431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., Zhang Y., Wang L. J. Biol. Chem. 2009;284:31921–31927. doi: 10.1074/jbc.M109.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Yamashita T., Ishikawa M. Oncol. Rep. 2002;9:565–569. [PubMed] [Google Scholar]
- Martin M. D., Matrisian L. M. Cancer Metastasis Rev. 2007;26:717–724. doi: 10.1007/s10555-007-9089-4. [DOI] [PubMed] [Google Scholar]
- Coussens L. M., Tinkle C. L., Hanahan D., Werb Z. Cell. 2000;103:481–490. doi: 10.1016/s0092-8674(00)00139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang W., Lv Q., Zhu D. Oncol. Rep. 2015;33:3108–3116. doi: 10.3892/or.2015.3931. [DOI] [PubMed] [Google Scholar]
- Qin W., Dong P., Ma C., Mitchelson K., Deng T., Zhang L., Sun Y., Feng X., Ding Y., Lu X., He J., Wen H., Cheng J. Oncogene. 2012;31:4067–4075. doi: 10.1038/onc.2011.561. [DOI] [PubMed] [Google Scholar]
- Park S., Eom K., Kim J., Bang H., Wang H. Y., Ahn S., Kim G., Jang H., Kim S., Lee D., Park K. H., Lee H. BMC Cancer. 2017;17:658. doi: 10.1186/s12885-017-3642-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V., Bell G. W., Nam J. W., Bartel D. P. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei W.-J., Liu J., Chao H., Ji L.-N., Li A.-X., Liu J.-Z. Transition Met. Chem. 2003;28:852–857. [Google Scholar]
- Marković V., Janićijević A., Stanojković T., KolundŽija B., Sladić D., Vujčić M., Janović B., Joksović L., Djurdjević P. T., Todorović N., Trifunović S., Joksović M. D. Eur. J. Med. Chem. 2013;64:228–238. doi: 10.1016/j.ejmech.2013.03.071. [DOI] [PubMed] [Google Scholar]
- Deshpande M. S., Kumbhar A. A., Kumbhar A. S., Kumbhakar M., Pal H., Sonawane U. B., Joshi R. R. Bioconjugate Chem. 2009;20:447–459. doi: 10.1021/bc800298t. [DOI] [PubMed] [Google Scholar]
- Pyle A. M., Rehmann J. P., Meshoyrer R., Kumar C. V., Turro N. J., Barton J. K. J. Am. Chem. Soc. 1989;111:3051–3058. [Google Scholar]
- Long E. C., Barton J. K. Acc. Chem. Res. 1990;23:271–273. [Google Scholar]
- Shi J.-H., Zhou K.-L., Lou Y.-Y., Pan D.-Q. Spectrochim. Acta, Part A. 2018;193:14–22. doi: 10.1016/j.saa.2017.11.061. [DOI] [PubMed] [Google Scholar]
- Ahmad A., Ahmad M. Spectrochim. Acta, Part A. 2018;188:244–251. doi: 10.1016/j.saa.2017.07.014. [DOI] [PubMed] [Google Scholar]
- Kakkar R., Garg R., Suruchi J. Mol. Struct.: THEOCHEM. 2002;584:37–44. [Google Scholar]
- Azqueta A., Collins A. R. Arch. Toxicol. 2013;87:949–968. doi: 10.1007/s00204-013-1070-0. [DOI] [PubMed] [Google Scholar]
- Kim J. M., Kim S. K. Bull. Korean Chem. Soc. 2011;32:964–972. [Google Scholar]
- Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Ohno M., Abe T. J. Immunol. Methods. 1991;145:199–203. doi: 10.1016/0022-1759(91)90327-c. [DOI] [PubMed] [Google Scholar]
- Kontogiorgis C., Hadjipavlou-Litina D. J. Enzyme Inhib. Med. Chem. 2003;18:63–69. doi: 10.1080/1475636031000069291. [DOI] [PubMed] [Google Scholar]
- Mihailović N., Marković V., Matić I. Z., Stanisavljević N. S., Jovanović ž. S., Trifunović S., Joksović Lj. RSC Adv. 2017;7:8550–8560. [Google Scholar]
- Ormerod M. G., in Flow cytometry, A practical approach, Oxford University Press, 2000. [Google Scholar]
- Aranda E., Owen G. I. Biol. Res. 2009;42:377–389. [PubMed] [Google Scholar]
- Matić I. Z., Aljančić I., Vajs V., Jadranin M., Gligorijević N., Milosavljević S., Juranić Z. D. Nat. Prod. Commun. 2013;8:1291–1296. [PubMed] [Google Scholar]
- Reichmann M. E., Rice S. A., Thomas C. A., Doty P. J. Am. Chem. Soc. 1954;76:3047–3053. [Google Scholar]
- Vijayalakshmi R., Kanthimathi M., Subramanian V., Nair B. U. Biochem. Biophys. Res. Commun. 2000;271:731–734. doi: 10.1006/bbrc.2000.2707. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Weber G. Biochemistry. 1973;12:4161–4170. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F. and Maniatis T. in Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, USA, 2nd edn, 1989. [Google Scholar]
- Janović B. S., Collins A. R., Vujčić Z. M., Vujčić M. T. J. Hazard. Mater. 2017;321:576–585. doi: 10.1016/j.jhazmat.2016.09.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.