Figure 3.
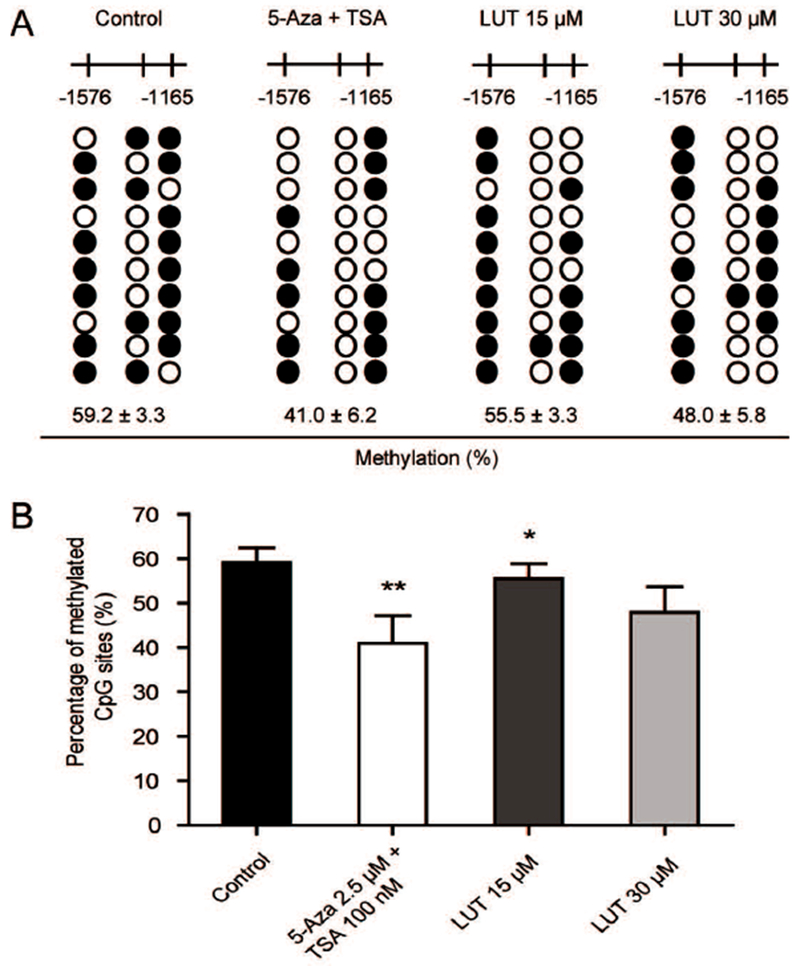
LUT reduced the CpG methylation ratio of the Nrf2 promoter region. (A-B) Detailed methylation patterns of 3 selected CpG sites (−1576 to −1165) in the promoter region of the Nrf2 gene in HCT116 cells were determined by bisulfite genomic sequencing. Filled dots indicate the methylated CpG sites, and open circles indicate the unmethylated CpG sites. HCT116 cells were treated with various concentrations of LUT or a positive control, 5-Aza and TSA, for 5 days, and genomic DNA was then bisulfite converted. Methylated cytosine was determined by Sanger sequencing. Ten clones were selected to represent three independent experiments. The methylation percentage was calculated from three independent experiments as the number of methylated CpG sites over the total number of CpG sites examined. The data are presented as the mean ± SD. * P < 0.05 and ** P < 0.01 versus the control group.
