Figure 1.
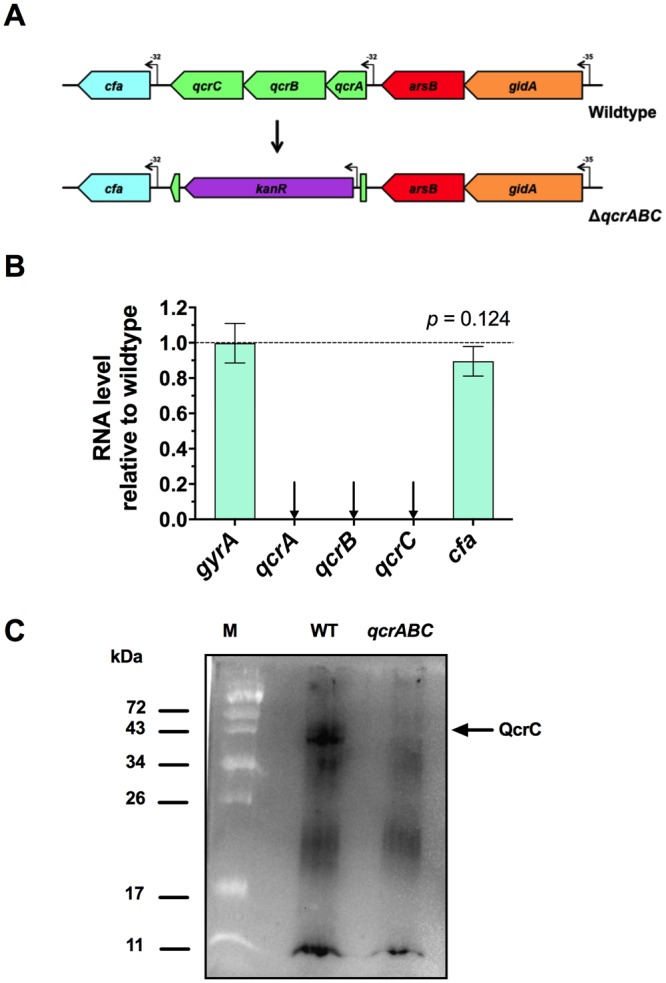
Construction and verification of a qcrABC deletion mutant. (A) Mutagenesis strategy. The majority of the coding regions of the qcrA-C genes were replaced with a kanamycin resistance cassette with its own promoter. The downstream cfa gene has its own promoter, but the cassette has no terminator and so should be non-polar on cfa. (B) RT-PCR to verify expression of cfa gene and absence of qcr gene transcription in the qcrABC mutant. Fold-change in the mutant is shown relative to the wild type strain, using gyrA gene as a control. The difference between expression of the cfa gene in mutant and wild-type was not significant by t-test. (C) shows a comparison of the membrane associated c-type cytochromes revealed after a total membrane preparation was subjected to SDS-PAGE and electroblotting to a nitrocellulose membrane, followed by staining for haem-associated peroxidase activity using the chemiluminescence technique described in Methods. 20 μg total protein was run per lane. The Image shown is a cropped version of the full-size blot that can viewed in Supplementary Fig. 1, and was obtained using a ChemiDoc XRS system (BioRad Inc) with an exposure time of 2 min. A band corresponding to the expected size of QcrC is missing in the qcrABC deletion mutant.
