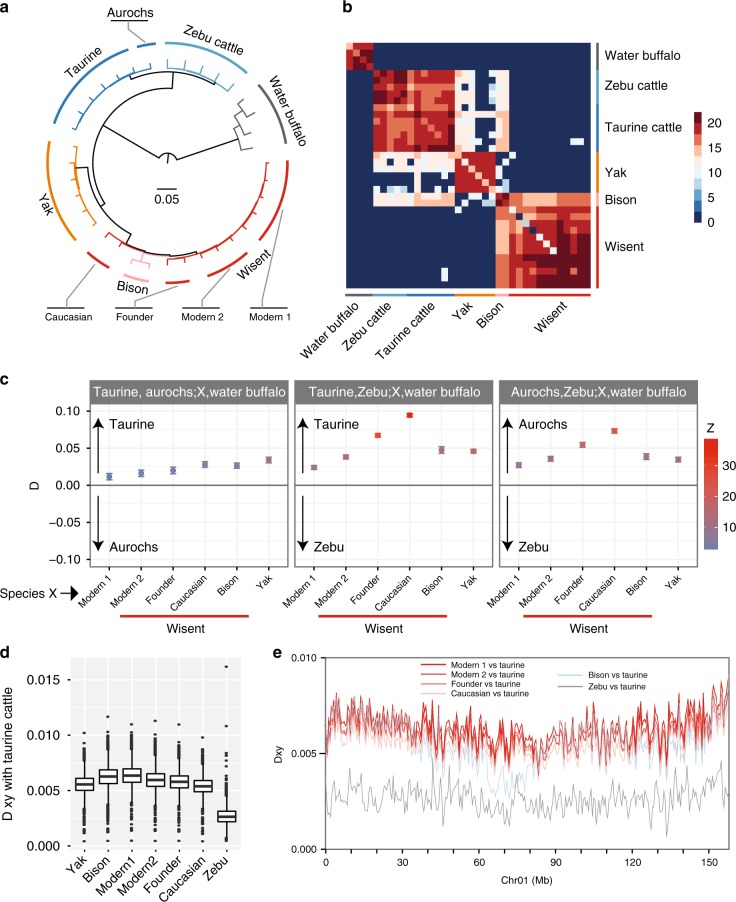
Fig. 2.
Phylogenetic relationships and inference of gene flow between six species. a Tree of allele-sharing distances between 36 individuals based on SNP calling. b Log total length of identical-by-descent haplotypes shared by individuals, the dark red corresponding to e20 = 454 Mbp. c ABBA/BABA statistic D with the indicated populations. All D values shown are significant (Z-score > 3, and p value < 0.00135). Positive values indicate that species X is more related to the first species than to the second species (in the first panel taurine cattle and aurochs, respectively). The respective wisent populations differ in their degree of gene flow with cattle lineages. d Pairwise genome-wide Dxy genetic distances between taurine cattle and zebu cattle, yak, American bison, and other wisent populations. X indicates the respective species on the X-axis. e Dxy distances between taurine cattle and zebu, different wisent populations, or American bison along chromosome 1. The Caucasian wisent population has the lowest Dxy in most regions, but not in the regions from 60 to 80 Mb; American bison is remarkably close to taurine cattle, which is not observed in other chromosomes