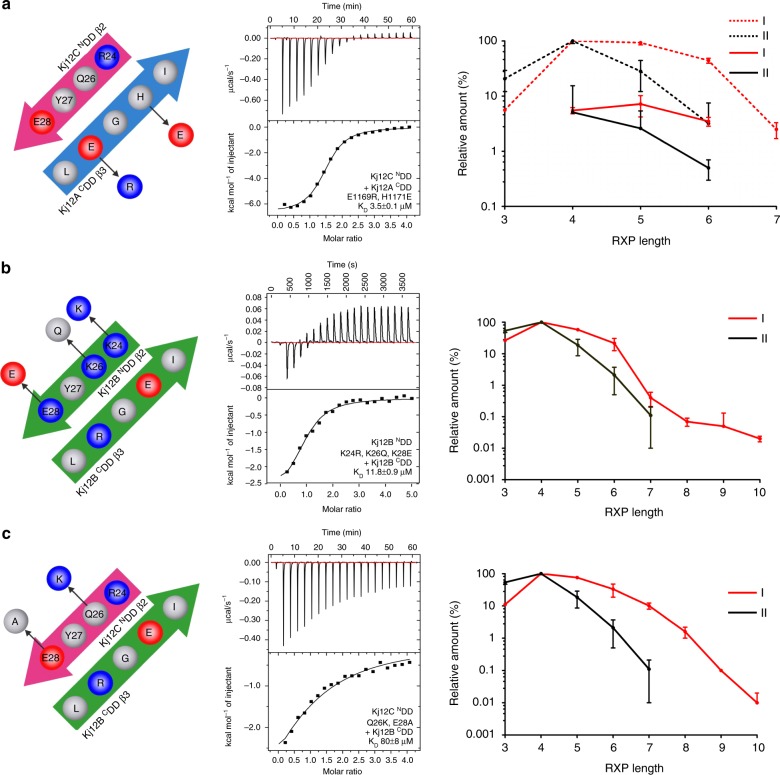
Fig. 5.
Optimization of CDD or NDDs in Kj12ABC system for the production of longer RXPs. Additionally, the ITC thermograms and the derived binding curves for titrations between optimized CDD or NDDs variants are shown. a Kj12A-CDD was optimized by two amino acid exchanges, E1169R and H1171E, on Kj12A-CDD. Co-expression of natural Kj12B with optimized Kj12C led to increased production of longer RXPs. Red lines (I) represent RXP production in the modified system, black lines (II) represent RXP production in the natural Kj12BC. Solid lines indicate fully methylated Val (mV) RXPs, dashed lines indicate RXPs containing only one non-methylated Val. b Optimization of Kj12B-NDD for tighter interaction with Kj12B-CDD via three amino acid exchanges, K26Q, K24R and K28E, on Kj12B-NDD. A red line (I) represents RXP production in the optimized system, a black line (II) represents RXP production in the natural Kj12BC. Only fully methylated RXPs are shown. c In addition to amino acid exchanges in b, Kj12C-NDD was additionally modified via two amino acid exchanges, Q26K and E28A, to reduce the interaction with Kj12B-CDD and allowing better interaction between Kj12B-NDD and Kj12B-CDD. No RXPs were detected after co-expression of natural Kj12B and modified Kj12C-NDD probably due to very weak affinities. A red line (I) represents RXP production in the optimized system, a black line (II) represents RXP production in natural Kj12BC. Only fully methylated RXPs are shown. x Axis, numbers of amino acid residues in RXPs (RXP length). y Axis, production of the corresponding RXPs relative to the most abundant derivative set to 100%