Abstract
Clindamycin is a protein synthesis inhibitory agent that has the ability to suppress the expression of virulence factors in Staphylococcus aureus. Recent guidelines recommend the use of clindamycin for the treatment of toxin-mediated infections. Clindamycin modulates virulence expression at sub-inhibitory concentrations (sub-MICs) in clindamycin-susceptible S. aureus strains but previous report shown that this effect was supressed for constitutive clindamycin resistant strains. However, no data are currently available on the impact of clindamycin at sub-MICs on the virulence of inducible clindamycin-resistant S. aureus strains. Here, we show that sub-MICs of clindamycin decrease Panton–Valentine leucocidin, toxic-shock-staphylococcal toxin (TSST-1) and alpha-haemolysin (Hla) expression in six inducible clindamycin-resistant isolates cultivated in vitro in CCY medium. These results suggest that the clindamycin anti-toxin effect is retained for inducible clindamycin-resistant S. aureus isolates; therefore, its usage should be considered within the treatment regimen of toxin related infections for inducible clindamycin-resistant S. aureus.
Keywords: Staphylococcus aureus, Panton–Valentine leucocidin, Toxic-shock-staphylococcal toxin, Alpha-haemolysin, Inducible clindamycin resistance, Anti-toxin effect
Introduction
Clindamycin is a protein synthesis inhibitory agent that has the ability to suppress the expression of virulence factors in Staphylococcus aureus at sub-inhibitory concentrations (sub-MICs). Indeed, several studies have reported the ability of clindamycin at sub-MICs to decrease the production of Panton-Valentine leucocidin (PVL), toxic-shock-staphylococcal toxin (TSST-1) or alpha-haemolysin (Hla) [1–6]. Therefore, recent guidelines recommend the use of clindamycin for the treatment of toxin-mediated infections (e.g., toxic shock syndrome and necrotizing pneumonia) [7]. This modulation of virulence expression by clindamycin occurs in clindamycin-susceptible S. aureus strains but is abolished in constitutive clindamycin-resistant strains [5]. Clindamycin resistance results from enzymatic methylation of the antibiotic binding site in the 50S ribosomal subunit (23S rRNA). The responsible methylase encoded by the erm gene can phenotypically result in a MLSB (macrolides, lincosamides, and group B streptogramins) resistance constitutive (in vitro resistance to all MLSB) or inducible (a positive “D-test” in agar diffusion method: resistance to erythromycin, susceptible to lincosamides with a flattening of the inhibition zone in regard to erythromycin disc) [8]. Contrary to constitutive clindamycin-resistant strains, no data are currently available on the impact of clindamycin at sub-MICs on the virulence of inducible clindamycin-resistant S. aureus strains. Here, we have shown in a selection of inducible clindamycin-resistant S. aureus strains that clindamycin maintains its anti-toxin effect at sub-inhibitory concentrations.
Methods
Bacterial strain selection
One hundred eighty S. aureus strains from the French National Reference Centre of Staphylococci were genotyped by the S. aureus Genotyping Kit 2.0 (Clondiag Alere® Jena, Germany), and the major macrolide resistance genes were recorded (ermA/B/C/T, linA, msrA, vatA/B, vgaA, vgb, and mbpB). The mecA gene responsible for methicillin resistance and the lukS-PVL and tst genes encoding for PVL and TSST-1, respectively, were also recorded.
Macrolide phenotypic resistance was determined by the agar diffusion disk assay using antibiotic discs (erythromycin, lincomycin, and quinupristin-dalfopristin) (I2A, Montpellier, France) and Mueller–Hinton E agar medium (bioMérieux, Marcy l’Etoile, France) according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations [9]. Inducible MLSB resistance was detected phenotypically by an inhibition zone between the erythromycin disk and the lincomycin disks indicating a positive D-test [8]. S. aureus strains exhibiting inducible MLSB resistance were selected, and the minimal inhibitory concentrations (MIC) of clindamycin (provided by Pfizer, Amboise, France) were determined by micro-dilution method using casein hydrolysate–yeast extract medium (CCY) broth medium. The S. aureus strain ATCC 29213 was used for clindamycin MIC determination in both CCY (homemade) and Mueller–Hinton (bioMérieux, Marcy-l’Etoile, France) broths.
Staphylococcus aureus culture conditions and toxin quantification
Since toxin production in Muller Hinton broth medium is very low, experiments were performed in CCY broth medium. Briefly, selected S. aureus strains were cultured overnight on Trypticase blood agar plates. Colonies were resuspended in CCY broth to a 0.5 McFarland adjusted turbidity. Cultures were performed at 37 °C with gyratory shaking (180 rpm). When the optical density reached a turbidity of a 2 McFarland (6 × 108 CFU/mL), clindamycin was added to the cultures to the final concentrations of 1/2 MIC, 1/4 MIC, and 1/8 MIC. Cultures with or without clindamycin (growth control) were incubated at 37 °C with shaking for 6 h (180 rpm) [3, 10]. After the incubation time, culture supernatants were collected by centrifugation at 10,000g for 15 min and used for toxin quantification. The PVL, Hla and TSST-1 levels in the supernatant were quantified using a specific ELISA assay as previously described [3], using ELISA kit provided by GlaxoSmithKline, Brentford, United Kingdom, and TSST-1 antibodies provided by Toxin technology, Sarasota, FL, USA. Percentages of toxin release for each condition were calculated related to growth control without antibiotic according to formula:
Statistical analyses
Statistical analyses were performed using RStudio, version 0.99.893 (RStudio Team (2009–2016)). RStudio: Integrated Development for R. RStudio, Inc., Boston, MA, USA). We performed t test to compare toxin expression under antibiotics conditions and growth control without antibiotics (= 100% of release).
Results
Inducible MLSB resistance in S. aureus strains and the MIC of clindamycin
Among the 180 strains tested, 78 were methicillin-susceptible S. aureus (MSSA), and 102 were methicillin-resistant S. aureus (MRSA). Among these 180 strains, 112 strains harboured at least one gene encoded macrolide resistance and 68 strains harboured none. Ninety-two strains harboured only one macrolide resistance gene: 28 strains harboured ermA, 2 strains harboured ermB, 40 strains harboured ermC, 9 strains harboured ermT (all 9 belonging to the clonal complex 398), 9 strains harboured vgaA, 3 strains harboured msrA and one strain harboured linA. Twenty strains contained two resistance genes: 17 strains contained msrA and mbpB, one strain contained ermC and linA, one strain contained ermC and vgaA and one strain contained msrA and vgaA.
Phenotypically, six strains tested positive for the D-test, indicating the presence of MLSB inducible clindamycin resistance (Table 1). Three of these strains were MRSA, including a European CA-MRSA (Community-acquired MRSA) clone, ST80 PVL+ (ST2015-0934), a variant ACME negative CA-MRSA USA300 clone, ST8 PVL+ (ST2015-0940) and a Geraldine MRSA clone, ST5 TSST-1+ (ST2015-1098). The three remaining S. aureus strains were MSSA, including a CC-49 PVL+ strain (ST2014-0018), a CC-30 TSST-1+ strain (ST2015-0773) and a CC-121 strain (ST2014-0942). The three MRSA and the three MSSA strains, respectively, carried the ermC and ermA genes responsible for MLSB inducible macrolide resistance. The MIC of clindamycin in CCY for six S. aureus strains was 0.25 mg/L, while the MIC of clindamycin for ATCC 29213 S. aureus was 0.06 mg/L in both CCY and Mueller–Hinton broth.
Table 1.
Inducible MLSB macrolide resistant strains used to explore clindamycin effect on virulence factors expression
| Strain reference | Description | Clone |
|---|---|---|
| ST2014-0018 | CC-49, mecA−, ermA+, PVL+ | – |
| ST2015-0773 | CC30, mecA−, ermA+, TSST-1 + | – |
| ST2014-0942 | CC121, mecA−, ermA+, PVL−, TSST-1- | – |
| ST2015-0934 | ST80, mecA+, ermC+, PVL+ | European CA-MRSA |
| ST2015-0940 | ST8, mecA+, ermC+, PVL+, ACME− | Variant USA 300 CA-MRSA |
| ST2015-1098 | ST5, mecA+, ermC+, TSST-1+ | Geraldine |
CA-MRSA community-acquired methicillin-resistant Staphylococcus aureus
Effect of clindamycin on Hla expression and release in MLSB inducible S. aureus strains
According to culture condition, means of Hla measurements (two determinations) in growth control supernatant were equal to 60.96, 52.00, 39.84, 19.79, 3.23 and < 0.75 ng/mL for ST2014-0018, ST2015-0940, ST2015-0934, ST2014-0942, ST2015-1098 and ST2015-0773 respectively.
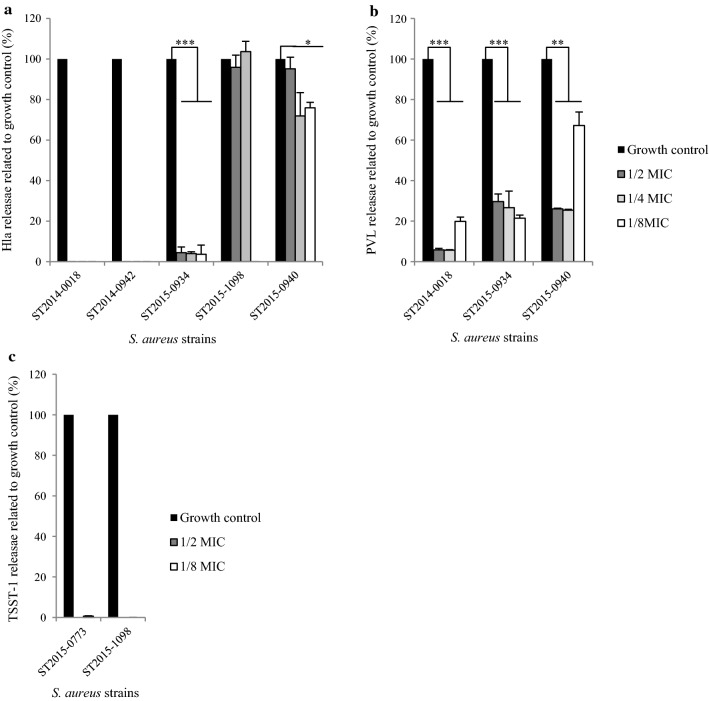
Sub-MICs of clindamycin (from 1/2 MIC to 1/8 MIC) induced a dramatic decrease of supernatant Hla for three strains (ST2014-0018, ST2014-0942 and ST2015-0934) ranging from undetectable Hla levels for ST2014-0018 and ST2014-0942 to low Hla levels (4.07%, CI95% 1.53, 6.62, p < 0.001) for ST2015-0934 at each clindamycin concentration (Fig. 1a). For ST2015-1098, only 1/8 MIC of clindamycin suppressed Hla production (undetectable level), and for ST2015-0940, we observed a slight significant decrease in Hla production (80.99%, CI95%, 67.81, 94.17, p < 0.05) at 1/4 MIC and 1/8 MIC of clindamycin (Fig. 1a). For the ST2015-0773 strain, Hla was undetectable by the ELISA assay regardless of the experimental conditions (control or with antibiotic) most likely due to a truncated Hla variant present in CC-30 strains, which the ELISA assay was unable to detect [11].
Fig. 1.
Virulence factors release by inducible MLSB-resistant S. aureus strains after sub-MIC of clindamycin treatment. Hla (a), PVL (b) and TSST-1 (c) release were measured by ELISA detection after clindamycin treatment (1/2 MIC, 1/4 MIC and 1/8 MIC) in comparison to the growth control without clindamycin. The averages represent two determinations for each condition. For each strain, virulence factor release at 1/2 MIC, 1/4 MIC and 1/8 MIC of clindamycin were pooled (n = 6) to perform the t test to compare with growth control without antibiotic (= 100%) and calculate the 95% confidence interval of the average. *p < 0.05; **p < 0.01; ***p < 0.001. Lack of a chart bar means that the factor virulence release measure was below the detection limit (i.e., 0.75 ng/mL for Hla and 1.56 ng/mL for TSST-1)
Effect of clindamycin on PVL expression and release in MLSB inducible S. aureus strains
According to culture condition, means of PVL measurements (two determinations) in growth control supernatant were equal to 682.43, 327.30 and 161.89 ng/mL for ST2015-0940, ST2015-0934 and ST2017-0018 respectively.
Of the six S. aureus strains tested, three stains carried the lukS-PVL gene. For these three strains, the sub-MICs of clindamycin significantly decreased PVL release compared to that of the growth control (without antibiotics) (Fig. 1b). PVL concentrations, expressed as percentages of the growth control levels, in the supernatants treated with clindamycin at sub-MICs ranged from 10.46% (CI95%, 2.74, 18.18, p < 0.01) for ST2014-0018, 25.93% (CI95%, 20.15, 31.71, p < 0.001) for ST2015-0934, and 39.55% (CI95%, 16.87, 62.23, p < 0.01) for ST2015-0940.
Effect of clindamycin on TSST-1 expression and release in MLSB inducible S. aureus strains
Of the six S. aureus strains tested, two carried the tst gene and exhibited variable TSST-1 expression in comparison to that of the growth control: 251.04 ng/mL for ST2015-0773 and 2.49 ng/mL for ST2015-1098. The sub-MICs of clindamycin induced a dramatic decrease of TSST-1 release for ST2015-0773 with an undetectable TSST-1 expression at 1/2 MIC of clindamycin and an approximate 99% decrease at 1/8 MIC. For ST2015-1098, 1/2 MIC and 1/8 MIC of clindamycin resulted in an undetectable TSST-1 expression level (Fig. 1c).
Discussion and conclusions
Staphylococcus aureus produces many virulence factors that play an important role in the pathogenesis of infection, such as Hla [12], PVL [13, 14] and TSST-1 [15]. Several studies have shown that clindamycin displays an anti-toxin in vitro effect at sub-inhibitory concentrations, inducing a decrease in toxin release [1–6]. Moreover, in a rabbit model of PVL+ CA-MRSA-induced necrotizing pneumonia, clindamycin was superior to vancomycin in reducing PVL induced tissue damage and the overall mortality rate, consistent with decreased PVL pulmonary concentrations [16]. Several case reports of necrotizing pneumonia and staphylococcal toxic shock highlight clindamycin’s efficacy when used as an adjunctive antibiotic for the treatment of S. aureus related toxin infections [17]. Notably, a retrospective analysis of 92 cases of CA-MRSA necrotizing pneumonia, mainly due to PVL-producing strains, showed improved clinical outcomes in patients treated with antimicrobials inhibiting toxin production (linezolid or clindamycin) [18]. The underlying mechanism of clindamycin’s anti-toxin effect is linked to the ribosome-blocking action in which the transcription of exoprotein and SaeRS global regulator system, are reduced [5, 19]. This anti-toxin effect is abolished by the constitutive MLSB resistance mechanism [5] in macrolide and lincosamide-resistant strains.
Clindamycin resistance can be constitutive or inducible. S. aureus strains belonging to the latter category display, when tested, in vitro susceptibility to clindamycin. Nevertheless, in the first decade of the 21st century, treatment failure was reported in adult and paediatric populations when clindamycin was used for inducible MLSB S. aureus infections [8, 20]. Consequently, the Clinical and Laboratory Standards Institute and EUCAST recommend searching and reporting for the inducible MLSB phenotype, but also stating that clindamycin may still be used for short-term therapy [9, 20].
Nevertheless, no data are currently available on the anti-toxin effect of clindamycin in inducible MLSB-resistant S. aureus isolates. Here, we showed that sub-MICs of clindamycin efficiently resulted in a decrease in PVL, Hla and TSST-1 production for inducible MLSB S. aureus strains in vitro, regardless of the methicillin susceptibility. For one strain, ST2015-1098, displaying a very weak level of Hla expression in control condition (3.23 ng/mL), we detected suppressed Hla production only 1/8 MIC of clindamycin. We suspected a lack of sensitivity of the measure method which could have failed to detect Hla decreased expression at 1/2 and 1/4 MIC of clindamycin, explaining the discrepancies between antibiotics concentrations. Interestingly, we observed that the decrease of virulence factor release (Hla and PVL) in the presence of clindamycin at sub-MICs was less altered for the variant USA300 clone (ST2015-0940). We hypothesize that this effect is likely dependent on the genetic background. This observation is in agreement with the study of Martin-Cardot et al., which had shown that the USA300 clone was resistant to staphylococcal protein A expression modulation by antibiotics and antimicrobial peptides [10].
In this short report, we chose to focus on the phenotypic expression of toxins, with possible direct impact on the clinical management of infections related to staphylococcal toxins, and without transcriptomic data because we and other authors, previously observed that modification at transcriptomic level did not always reflect modulation of toxins production [17].
In conclusion, our results indicate that the anti-toxin effect of clindamycin should therefore be considered when treating toxin related infections for inducible clindamycin-resistant S. aureus. However, given the risk of constitutive MLSB resistance acquisition upon clindamycin treatment, clindamycin should be coupled with a bactericidal effective antibiotic for the treatment of toxin-related S. aureus severe infections.
Authors’ contributions
EH, GL and OD designed the experiments. EH, CeB, CaB, MB and AT performed the experiments and collected data. EH analyzed data collection. EH, FV, GL and OD wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank the ECCMID (European Congress of Clinical Microbiology and Infectious Diseases) 2017 Programme Committee to have accepted this work for ECCMID 2017 congress (paper poster session).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CA-MRSA
community-acquired methicillin-resistant Staphylococcus aureus
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- Hla
alpha-haemolysin
- MIC
minimal inhibitory concentration
- MLSB
macrolides, lincosamides, and group B streptogramins
- MRSA
methicillin-resistant Staphylococcus aureus
- MSSA
methicillin-susceptible Staphylococcus aureus
- PVL
Panton–Valentine leucocidin
- Sub-MICs
sub-inhibitory concentrations
- TSST-1
toxic-shock-staphylococcal toxin
Contributor Information
Elisabeth Hodille, Phone: +334 72 07 16 90, Email: elisabeth.hodille@chu-lyon.fr.
Cédric Badiou, Email: cedric.badiou@univ-lyon1.fr.
Caroline Bouveyron, Email: caroline.bouveyron@chu-lyon.fr.
Michèle Bes, Email: michele.bes@chu-lyon.fr.
Anne Tristan, Email: anne.tristan@chu-lyon.fr.
François Vandenesch, Email: francois.vandenesch@chu-lyon.fr.
Gérard Lina, Email: gerard.lina@chu-lyon.fr.
Oana Dumitrescu, Email: oana.dumitrescu@chu-lyon.fr.
References
- 1.Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, Bryant AE. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis. 2007;195:202–211. doi: 10.1086/510396. [DOI] [PubMed] [Google Scholar]
- 2.Dumitrescu O, Boisset S, Badiou C, Bes M, Benito Y, Reverdy M-E, et al. Effect of antibiotics on Staphylococcus aureus producing Panton–Valentine leukocidin. Antimicrob Agents Chemother. 2007;51:1515–1519. doi: 10.1128/AAC.01201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otto MP, Martin E, Badiou C, Lebrun S, Bes M, Vandenesch F, et al. Effects of subinhibitory concentrations of antibiotics on virulence factor expression by community-acquired methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2013;68:1524–1532. doi: 10.1093/jac/dkt073. [DOI] [PubMed] [Google Scholar]
- 4.Ohlsen K, Ziebuhr W, Koller KP, Hell W, Wichelhaus TA, Hacker J. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother. 1998;42:2817–2823. doi: 10.1128/AAC.42.11.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbert S, Barry P, Novick RP. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect Immun. 2001;69:2996–3003. doi: 10.1128/IAI.69.5.2996-3003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickgiesser N, Wallach U. Toxic shock syndrome toxin-1 (TSST-1): influence of its production by subinhibitory antibiotic concentrations. Infection. 1987;15:351–353. doi: 10.1007/BF01647737. [DOI] [PubMed] [Google Scholar]
- 7.Nathwani D, Morgan M, Masterton RG, Dryden M, Cookson BD, French G, et al. Guidelines for UK practice for the diagnosis and management of methicillin-resistant Staphylococcus aureus (MRSA) infections presenting in the community. J Antimicrob Chemother. 2008;61:976–994. doi: 10.1093/jac/dkn096. [DOI] [PubMed] [Google Scholar]
- 8.Lewis JS, Jorgensen JH. Inducible clindamycin resistance in Staphylococci: should clinicians and microbiologists be concerned? Clin Infect Dis Off Publ Infect Dis Soc Am. 2005;40:280–285. doi: 10.1086/426894. [DOI] [PubMed] [Google Scholar]
- 9.v_8.0_Breakpoint_Tables.pdf. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf. Accessed 10 Jan 2018.
- 10.Cardot Martin E, Michel A, Raynal B, Badiou C, Laurent F, Vandenesch F, et al. Community-acquired meticillin-resistant Staphylococcus aureus strain USA300 resists staphylococcal protein A modulation by antibiotics and antimicrobial peptides. Int J Antimicrob Agents. 2015;45:19–24. doi: 10.1016/j.ijantimicag.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Xiao M, Zhao R, Zhang Q, Fan X, O’Sullivan MVN, Li D-F, et al. Genotypic diversity of Staphylococcus aureus α-hemolysin gene (hla) and its association with clonal background: implications for vaccine development. PLoS ONE. 2016;11:e0149112. doi: 10.1371/journal.pone.0149112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berube BJ, Bubeck Wardenburg J. Staphylococcus aureus α-toxin: nearly a century of intrigue. Toxins. 2013;5:1140–1166. doi: 10.3390/toxins5061140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillet Y, Issartel B, Vanhems P, Fournet J-C, Lina G, Bes M, et al. Association between Staphylococcus aureus strains carrying gene for Panton–Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–759. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 14.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton–Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis Off Publ Infect Dis Soc Am. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 15.Todd J, Fishaut M, Kapral F, Welch T. Toxic-shock syndrome associated with phage-group-I Staphylococci. Lancet Lond Engl. 1978;2:1116–1118. doi: 10.1016/S0140-6736(78)92274-2. [DOI] [PubMed] [Google Scholar]
- 16.Croisier-Bertin D, Hayez D, Da Silva S, Labrousse D, Biek D, Badiou C, et al. In vivo efficacy of ceftaroline fosamil in a methicillin-resistant Panton–Valentine leukocidin-producing Staphylococcus aureus rabbit pneumonia model. Antimicrob Agents Chemother. 2014;58:1855–1861. doi: 10.1128/AAC.01707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodille E, Rose W, Diep BA, Goutelle S, Lina G, Dumitrescu O. The role of antibiotics in modulating virulence in Staphylococcus aureus. Clin Microbiol Rev. 2017;30:887–917. doi: 10.1128/CMR.00120-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouzic N, Janvier F, Libert N, Javouhey E, Lina G, Nizou J-Y, et al. Prompt and successful toxin-targeting treatment of three patients with necrotizing pneumonia due to Staphylococcus aureus strains carrying the Panton–Valentine leukocidin genes. J Clin Microbiol. 2010;48:1952–1955. doi: 10.1128/JCM.01892-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiol Read Engl. 2003;149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- 20.Woods CR. Macrolide-inducible resistance to clindamycin and the D-test. Pediatr Infect Dis J. 2009;28:1115–1118. doi: 10.1097/INF.0b013e3181c35cc5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.