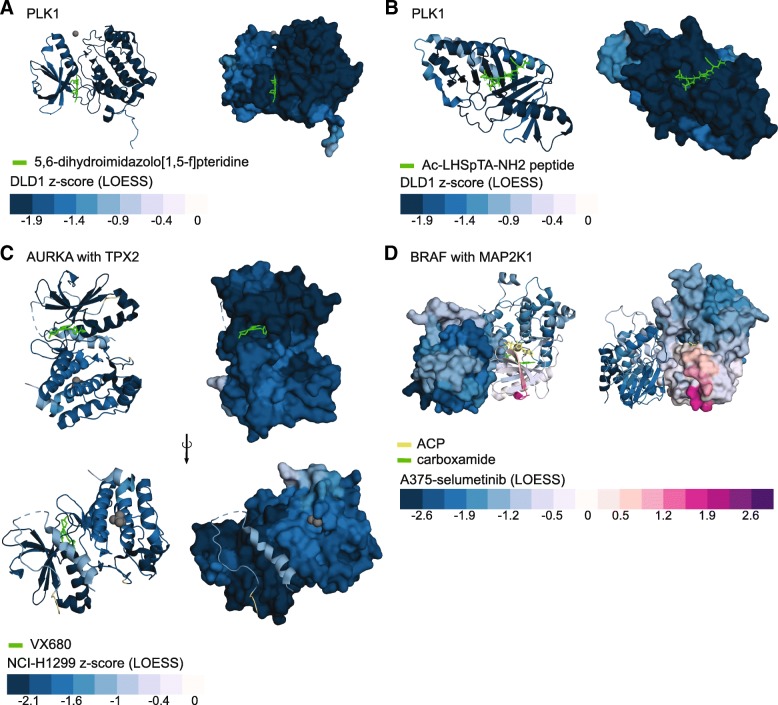
Fig. 4.
CRISPRO 3D structure maps. a PLK1, PDB ID: 5TA6. Mapped scores are DLD1 z-score (LOESS interpolation) of PLK1 (protein kinase domain, AA37-330, cartoon presentation in the left panel, surface presentation in the right panel) in complex with 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor (green). Zinc ion is displayed as a gray sphere. b PLK1, PDB ID 3FVH. Mapped scores are DLD1 z-score (LOESS interpolation) of PLK1 (polo box domain, AA368-604) in complex with Ac-LHSpTA-NH2 peptide. Both surface (right) and cartoon (left) presentation shown. C) AURKA with TPX2, PDB ID 3E5A. Mapped scores are NCI-H1299 z-score (LOESS interpolation) of AURKA (presented as surface in left panels, right as a cartoon, AA125-389, protein kinase domain) and TPX2 (presented solely as cartoon, AAs 6–21, 26–42, Aurora-A binding domain) in complex with VX680, an ATP-competitive small molecule inhibitor. Sulfate ions are displayed as gray spheres. d BRAF and MAP2K1, PDB ID 4MNE. Mapped scores A375 selumetinib (LOESS interpolation) of BRAF (surface in left panel, cartoon in right, AAs 449–464, 469–722, protein kinase domain) and MAP2K1 (cartoon in left panel, surface in right, AAs 62–274, 307–382, protein kinase domain). Ligands ACP in yellow, and 7-fluoro-3-[(2-fluoro-4-iodophenyl)amino]-N-{[(2S)-2-hydroxypropyl]oxy}furo[3,2-c]pyridine-2-carboxamide in green. Magnesium ion is displayed as a gray sphere