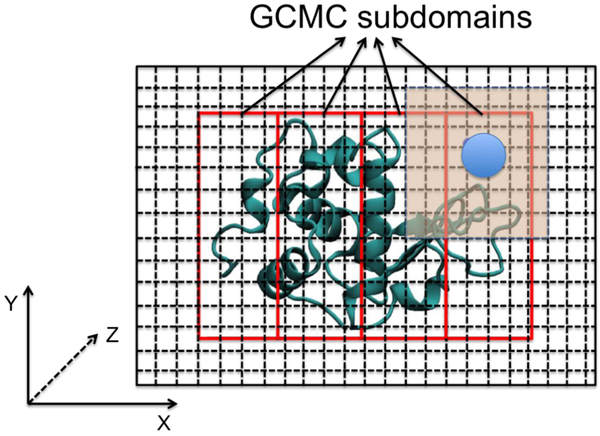
Figure 1.
In the GCMC simulation program, the three-dimensional simulation box is compartmentalized into a large number of small cubes with a length of 1 Å. All atoms in the simulation box are assigned into the cubes based on the atomic coordinates and the cube positions. By constructing a two-dimensional cube-atom array, all neighboring atoms surrounding an ion within a cutoff distance can be quickly found. In addition, a GCMC region can be defined if the simulation system contains biomolecules like protein or DNA. For parallel GCMC simulations, the GCMC region is divided into a number of GCMC subdomains.