Fig. 2.
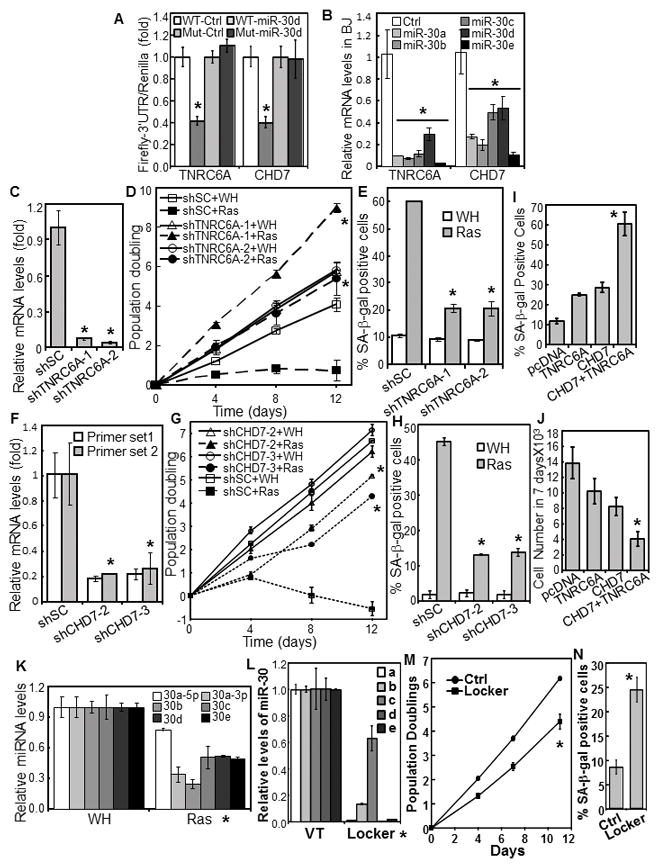
miR-30 disrupts senescence by targeting TNRC6A and CHD7.
(A) miR-30 suppresses the 3′UTR-reporters of TNRC6A and CHD7. The 3′UTR Firefly luciferase reporters of TNRC6A and CHD7 containing wild type (WT) or mutant (Mut) miR-30-binding sites were cotransfected into 293 cells with CMV-Renilla Luciferase and miR-30d or vector (Ctrl). Firefly luciferase activity was determined 24 hrs later and normalized to Renilla luciferase activity.
(B) TNRC6A and CHD7 mRNA were determined in BJ cells transduced with miR-30 members or vector (Ctrl) by qRT-PCR.
(C, F) TNRC6A (C) or CHD7 (F) mRNA were measured in BJ cells transduced with shRNA for TNRC6A (C) or CHD7 (F) or a scrambled shRNA (shSC) by qRT-PCR.
(D, G) Growth curves of BJ cells transduced with shRNA for TNRC6A (D) or CHD7 (G) or scrambled shRNA (shSC) and HaRasV12 (Ras) or vector (WH).
(E, H) % of SA-β-gal-positive cells in populations in (D) and (G), respectively, on day-9 of the growth curves.
(I–J) BJ cells transduced with miR-30c and HaRasV12 were transfected with TNRC6A, CHD7, both or vector (pcDNA3) together with pcDNA3-GFP. GFP-positive cells were sorted and stained for SA-β-gal (I) and numbers counted after 7 days (J).
(K) Levels of miR-30 family members were measured in BJ cells transduced with HaRasV12 (Ras) or vector (WH) by qRT-PCR.
(L–M) Inhibition of miR-30 induces senescence. BJ cells transduced with a miR-30-Locker that inhibits the expression of all miR-30 isoforms (Locker, Biosettia) or vector control (Ctrl) were analyzed for the expression levels of miR-30a–e by qRT-PCR (L), and oncogenic ras-induced senescence by growth curve analysis (M) and SA-β-gal staining (N).
(A–N) Values are mean±SD for triplicates. * p < 0.05 vs Ctrl, shSC, WH, or VT by Student t test.
