Fig. 5.
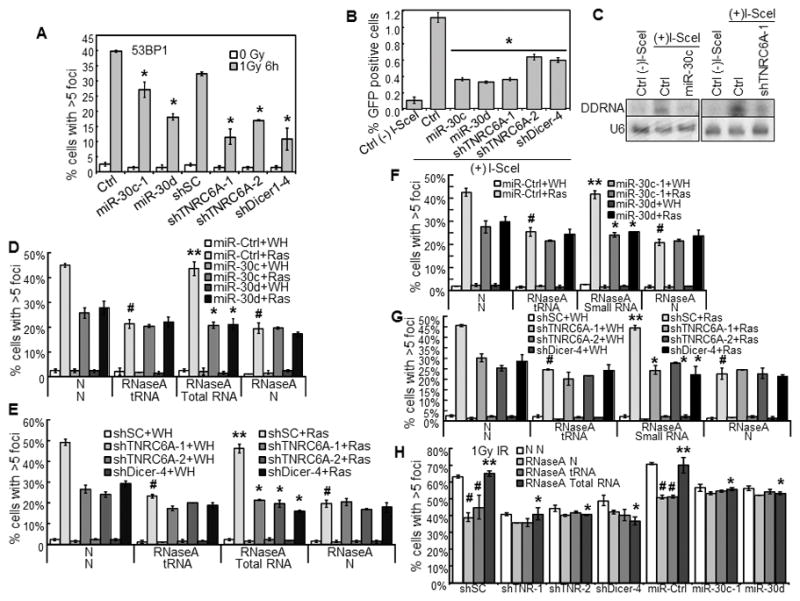
miR-30 and TNRC6A shRNA disrupt γ-radiation induced 53BP1-foci, HR-mediated repair, and expression and functionality of DDRNAs.
(A) % of cells with 53BP1-foci in BJ cells transduced with miR-30 or vector (Ctrl) or shRNA for TNRC6A or Dicer or a scrambled sequence (shSC) and treated with 0 or 1Gy γ-radiation.
(B) miR-30 and TNRC6A shRNA disrupt HR-mediated repair. I-SceI-EGFP-U2OS cells were transduced with miR-30, shRNA for TNRC6A or Dicer or vector control (Ctrl), and then transduced with I-SceI [(+)I-SceI] or control [(−)I-SceI]. % of GPF positive cells was determined by flow cytometry 7 days later.
(A, B) * p < 0.05 vs Ctrl or shSC by Student t test.
(C) miR-30 and TNRC6A shRNA reduce the DDRNA levels. Autoradiogram of Northern blot analysis of DDRNAs in I-SceI-EGFP-U2OS cells after transduction with miR-30c-1, shRNA for TNRC6A or vector (Ctrl) and I-SceI [(+)I-SceI] or control [(−)I-SceI]. A radio-labeled DNA fragment from the region surrounding the I-SceI site was used as probe for DDRNAs. U6 RNA was used as loading control.
(D–H) miR-30 and TNRC6A shRNA abrogate the functionality of DDRNA. BJ cells transduced with miR-30 or vector (miR-Ctrl) (D, F) or shRNA for TNRC6A or Dicer or a scrambled sequence (shSC) (E, G) and then with HaRasV12 (Ras) or vector (WH) were permeabilized, treated with RNaseA or not (N), and after washing and incubation with RNasin, incubated with total (D–E) or small (≤200 nucleotides) (F–G) RNA isolated from senescent cells, yeast total RNA (tRNA) or vehicle (N). Cells were stained for 53BP1-foci and quantified.
(H) BJ cells transduced with shRNA for TNRC6A (shTNR-1, 2) or Dicer or a scrambled sequence (shSC) or miR-30 or vector (miR-Ctrl) were treated with 1Gy γ-radiation, permeabilized after 24 hrs, treated with RNaseA or not (N), and after washing and incubation with RNasin, incubated with total RNA isolated from 1Gy γ-radiation-treated BJ cells (total RNA), yeast total RNA (tRNA) or vehicle (N). Cells were stained for 53BP1-foci and quantified.
(A–B, D–H) Values are mean±SD for triplicates.
(D–H) # p < 0.05 vs N/N by Student t test. ** p < 0.05 vs RNaseA/tRNA by Student t test. * p < 0.05 vs miR-Ctrl+Ras (D, F) or shSC+Ras (E, G) or miR-Ctrl/shSC+IR (H) by Student t test.
