Fig. 4. DNA Methylation-Dependent Regulation of miR-30.
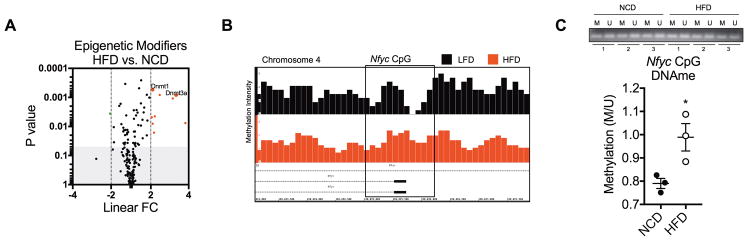
As shown in Figure 1, F4/80+ ATMs were isolated from epididymal fat of 22-week-old HFD-induced obese and lean mice. Gene expression and DNA methylation were evaluated in ATMs. (A) Volcano plot displaying linear FC of genes encoding epigenetic modification enzymes and factors. Fold change and p value observations were extracted from transcriptome microarrays (See Figure 1 and Figure S1). (B) IGB visualization of meDIP-seq peak intensity of DNA methylation in the Nfyc promoter CpG island. (C) Methylation-specific PCR quantification of DNA methylation (DNAme) in the Nfyc promoter CpG island. For (A and C), values presented are representative of 3 independent experiments with 20 pooled NCD mice and 10 pooled HFD mice per experiment. For (B), data are representative of one experiment of 60 pooled LFD and 30 pooled HFD mice. Statistical differences were determined by using Student’s t-test. *p<0.05.