SUMMARY
To permanently attach to surfaces, Caulobacter crescentus produces a strong adhesive, the holdfast. The timing of holdfast synthesis is developmentally regulated by cell cycle cues. When C. crescentus is grown in a complex medium, holdfast synthesis can also be stimulated by surface sensing, in which swarmer cells rapidly synthesize holdfast in direct response to surface contact. In contrast to growth in complex medium, here we show that when cells are grown in a defined medium, surface contact does not trigger holdfast synthesis. Moreover, we show that in a defined medium, flagellum synthesis and regulation of holdfast production are linked. In these conditions, mutants lacking a flagellum attach to surfaces over time more efficiently than either wild-type strains or strains harboring a paralyzed flagellum. Enhanced adhesion in mutants lacking flagellar components is due to premature holdfast synthesis during the cell cycle and is regulated by the holdfast synthesis inhibitor HfiA. hfiA transcription is reduced in flagellar mutants and this reduction is modulated by the diguanylate cyclase developmental regulator PleD. We also show that, in contrast to previous predictions, flagella are not necessarily required for C. crescentus surface sensing in the absence of flow, and that arrest of flagellar rotation does not stimulate holdfast synthesis. Rather, our data support a model in which flagellum assembly feeds back to control holdfast synthesis via HfiA expression in a c-di-GMP dependent manner under defined nutrient conditions.
Caulobacter crescentus produces a strong adhesive, the holdfast, to permanently attach to surfaces and form biofilms. In this study, we show that flagellum assembly is developmentally connected to the production of holdfast dependent upon both nutrient availability and c-di-GMP. These results provide new insight into the regulation of single cell adhesion and initial surface colonization leading to the process of biofilm formation.
INTRODUCTION
In natural environments, microorganisms form complex multicellular surface-associated communities known as biofilms. The first step of biofilm formation is the reversible attachment of single cells to the surface (Goller & Romeo, 2008, Tolker-Nielsen, 2015, Berne et al., 2018). For most bacteria, the initial attachment step requires flagellar motility and pili. These extracellular appendages presumably help cells overcome the hydrodynamic and electrostatic forces encountered close to surfaces: motility provides the necessary force to penetrate the repulsive barrier, and the flagellum and pili reduce the radius of electrostatic repulsion between the cell and the surface by initiating the first contact with the surface (Donlan, 2002, Berne et al., 2018). Once single cells have reversibly attached to the surface, irreversible attachment is mediated by strong adhesins, including adhesive proteins and extracellular polysaccharides (Karatan & Watnick, 2009, Fong & Yildiz, 2015, Berne et al., 2015).
C. crescentus is a robust model organism for studying initial attachment because of its ability to rapidly synthesize the adhesive holdfast in direct response to surface contact (Li et al., 2012, Hoffman et al., 2015, Ellison et al., 2017). C. crescentus has a dimorphic lifestyle: at each division, two different cell types are produced, a motile swarmer cell and a sessile stalked cell (Fig. 1A). The swarmer cell harbors a polar flagellum and several pili at the same pole and is unable to replicate DNA. The swarmer cell differentiates into a replication-competent stalked cell by shedding its flagellum, retracting its pili, and synthesizing a holdfast and a stalk at the same pole of the cell. The holdfast mediates permanent attachment (Ong et al., 1990) and is synthesized by a Wzy-dependent pathway similar to E. coli group 1 capsular polysaccharide biosynthesis (Smith et al., 2003, Toh et al., 2008, Hardy et al., 2010, Javens et al., 2013, Wan et al., 2013, Hardy et al., 2017).
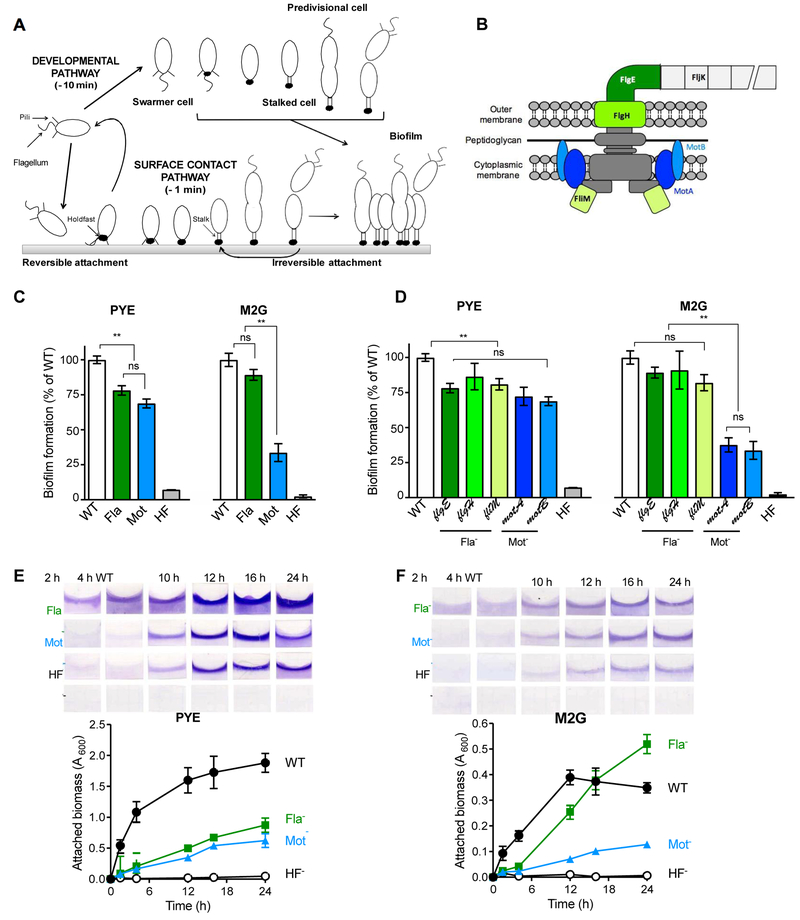
Fig. 1. Biofilm formation by Mot- and Fla- mutants under different nutrient conditions.
(A) Schematic representation of the C. crescentus life cycle as described in the text. Approximated timing of holdfast synthesis via the developmental and surface sensing pathways are given for WT in PYE, adapted from Table 1 and (Levi & Jenal, 2006, Ellison et al., 2017). (B) Schematic of the flagellum apparatus in C. crescentus, indicating the proteins relevant to this study. Mutations in proteins colored in green or blue result in Fla- or Mot- phenotypes respectively. (C, D) Biofilm formation in 24-well plates (after 16 h incubation at 30˚C) by (C) C. crescentus CB15 WT, ∆flgE, ∆motB and ∆hfsDAB mutant strains in PYE (Left) and M2G (Right); (D) different Fla- and Mot- mutants under the same conditions. Results are expressed as a percentage of WT biofilm formation. Error bars represent the standard error of the mean (SEM) of at least 3 independent replicates run in duplicate. Statistical comparisons are calculated using Student’s unpaired t-tests. ** P < 0.01; ns = not significant. (E-F) Amount of attached biomass quantified by crystal violet staining of biofilms grown on PVC coverslips standing vertically in 12-well at different time intervals in PYE (E) and M2G (F) media. Values are given as the average of triplicate samples of at least two independent experiments. The error is represented as SEM.
Holdfast synthesis is regulated at multiple levels. Transcription of both the holdfast synthesis (hfs) and attachment (hfa) genes is cell cycle regulated, and expression of both groups of genes peaks in predivisional cells. Similarly, transcription of the holdfast inhibitor gene, hfiA, is controlled by cell cycle developmental regulators (Fiebig et al., 2014). Upon division, swarmer cells are preloaded with a complete set of holdfast biosynthesis proteins as evidenced by the observation that neither RNA nor protein synthesis is required for holdfast synthesis in swarmer cells (Ellison et al., 2017, Levi & Jenal, 2006, Li et al., 2012). In these cells, HfiA prevents the holdfast synthesis machinery from producing holdfast prematurely by inhibiting HfsJ, a glycosyltransferase required for holdfast synthesis (Fiebig et al., 2014). Environmental factors such as blue-light via the LovK-LovR sensory system (Purcell et al., 2007) and nutrient availability via unknown regulators provide additional transcriptional control of hfiA (Fiebig et al., 2014). Thus, the probability of holdfast elaboration and population level attachment can be environmentally tuned via transcriptional control of hfiA.
In liquid cultures, holdfasts typically are not elaborated until the late swarmer stage (Levi & Jenal, 2006); however holdfast synthesis can be stimulated earlier in direct response to surface contact (Li et al., 2012, Hoffman et al., 2015, Ellison et al., 2017). In both cases, post-transcriptional signals activate holdfast biosynthesis. Increased c-di-GMP level during the swarmer-to-stalk transition is an important developmental trigger (Levi & Jenal, 2006, Abel et al., 2011, Sprecher et al., 2017). HfsJ, the glycosyltransferase targeted by HfiA, directly binds c-di-GMP and this binding stimulates holdfast synthesis (Hug et al., 2017). Additionally, degradation of HfiA during the swarmer phase may serve as a developmental trigger of holdfast elaboration.
The signal leading to surface stimulation of holdfast production has been linked to resistance to pilus retraction (Li et al., 2012, Ellison et al., 2017). Live microscopy imaging experiments show that swarmer cells contact the surface, stop rotating the flagellum in a pilus-dependent manner, and initiate holdfast synthesis (Li et al., 2012). We showed recently that physically blocking pilus retraction triggers holdfast synthesis in the absence of a surface (Ellison et al., 2017). In addition, c-di-GMP is produced by the diguanylate cyclase DgcB in response to a physical or chemical signal from the flagellar motor upon contact with a surface (Hug et al., 2017). This increase in c-di-GMP in turn stimulates HfsJ and holdfast production (Fiebig et al., 2014, Hug et al., 2017).
In this study, we investigate the role of the flagellum and its rotation in adhesion, holdfast synthesis and surface sensing of C. crescentus under different nutrient conditions. We compare mutants lacking the flagellum (Fla-) or the stator required for flagellum rotation (Mot-) to wild-type cells. Though both strains exhibit non-motile phenotypes, we identify several differences in adhesion between these strains that are conditional upon media composition. Both Fla- and Mot- mutants display similar attachment deficiencies in complex medium, while in defined medium, Fla- mutants form more robust biofilms over time, despite impaired initial adhesion. We find that the overall enhanced adhesion phenotype in a Fla- mutant in defined medium is due to an increase in the number of cells that synthesize holdfast, and a premature production of holdfast during the cell cycle. The initial attachment deficiency of Fla- and Mot- mutants in complex medium is not due to an inability to respond to surface contact. Moreover, we find that arrest of flagellar rotation does not stimulate holdfast synthesis, as previously proposed (Li et al., 2012). We conclude that neither the flagellum nor its rotation is required to sense surfaces by C. crescentus. Instead, our results show that the status of flagellum synthesis regulates holdfast synthesis by transcriptional control of the holdfast inhibitor gene hfiA via a signaling pathway that depends on the diguanylate cyclase PleD.
RESULTS
Mot- mutants are more impaired in biofilm formation than Fla- mutants in defined medium.
To dissect the role of motility and the flagellum in C. crescentus adhesion, we first constructed two types of non-motile mutants: (1) a strain lacking a flagellum (Fla-), by in frame deletion of the hook gene (∆flgE); and (2) a mutant harboring a paralyzed flagellum (Mot-), by in frame deletion of one of the flagellum stator genes (∆motB) (Fig. 1B). In addition, a mutant lacking the holdfast export genes, ∆hfsDAB, served as a holdfast-deficient (HF-) control. To assess long-term adhesion phenotypes of C. crescentus wild-type (WT) and mutant strains at the population level, cultures were first grown in complex peptone-yeast extract (PYE) medium under static conditions, and biofilm formation was measured by crystal violet staining (Fig. 1C). As expected, the HF- mutant was unable to form biofilms, as it lacks the ability to mediate permanent adhesion via the holdfast. Fla- and Mot- mutants exhibited similar deficiencies in biofilm formation in complex PYE medium (Fig. 1C), as previously reported (Bodenmiller et al., 2004, Levi & Jenal, 2006): both mutants formed approximately 25% less biofilm than WT after 24h incubation. We then performed the same type of experiment using defined M2G medium. In this condition, cells are provided sufficient inorganic phosphate, ammonium salts, and fixed carbon to support dense growth. However, they must synthesize de novo all organic building blocks such as amino acids and nucleotides. In this defined medium, growth rate is approximately two-fold slower than in complex PYE. We found that Fla- and Mot- mutants behaved differently in these nutrient conditions: Fla- mutants were less severely impaired than the Mot- mutant, forming around 10% and 65% less biofilm compared to WT, respectively (Fig. 1C). To determine if the medium-dependent differences in attachment between Fla- and Mot- mutants were consistent for other non-motile mutants, we examined biofilm formation in additional Fla- strains (outer membrane ring gene (∆flgH) and switch gene (fliM::Tn5)), and an additional Mot- (∆motA) mutant (Fig 1D). Again, while Mot- and Fla- mutants behaved in a similar manner in PYE medium (Fig. 1D), Mot- mutants were more severely impaired in biofilm formation in defined medium than Fla- mutants (Fig. 1D). Importantly, the motility and biofilm defect phenotypes of the Fla- ΔflgE and Mot- ΔmotB mutants can be complemented in trans by a replicating plasmid encoding a copy of flgE and motB, respectively (Fig. S1). Because there was no notable difference in biofilm formation between mutants within the Mot- or Fla- classes, we focused on the in-frame deletion ∆motB and ∆flgE mutants in all subsequent experiments.
To determine what stage of biofilm formation is impacted in the Mot- and Fla- mutants, we analyzed these mutants over time in a static system in PYE (Fig. 1E) and M2G (Fig. 1F). We quantified the amount of biomass attached on plastic coverslips standing in 12-well plates at different time intervals. In PYE medium, both Mot- and the Fla- were impaired in the first steps of adhesion (around 15% of biomass attached compared to WT for the first 4 h, Fig. 1E). These mutants achieved less than 50% of WT adhesion after 24 h of incubation (Fig. 1E). These results show that, when grown in PYE medium, the Mot- and the Fla- mutants are impaired in both initial attachment and biofilm maturation. However, we observed different behaviors between Mot- and Fla- mutants in M2G. First, both mutants formed less than 10% of attached biomass compared to the WT strain for the first 4 h (Fig. 1F), showing that both types of mutants are severely impaired in biofilm initiation. For the Mot- strain, this adhesion defect persisted through time. In contrast, while the Fla- mutant demonstrated poor initial adhesion, biofilm formation ultimately recovered to the same level as the WT strain after 16 h, and surpassed WT levels at later timepoints. These results indicate that, in M2G, motility is important for initial adhesion under static conditions, as both Mot- and Fla- mutants are deficient in the earliest stages of adhesion; however, lack of flagellar motor function remains detrimental for biofilm formation, while absence of a flagellum can be compensated for over time. (Fig. 1F). The phenotypic difference observed between the Fla- and Mot- mutants, both of which are non-motile, suggests that the regulation of biofilm formation in defined M2G medium is more complex than non-motility.
Surface-contact stimulation of holdfast synthesis is not impaired in Fla- and Mot- mutants.
The lack of motility in the mutants tested above cannot fully explain the biofilm defect, as we observed a different response for Mot- or Fla- mutants in defined growth medium. In C. crescentus, the most crucial component of irreversible adhesion to surfaces and biofilm formation is the holdfast (Ong et al., 1990, Bodenmiller et al., 2004, Entcheva-Dimitrov & Spormann, 2004). We thus sought to determine whether holdfast synthesis is misregulated in Mot- and Fla- mutants. C. crescentus holdfast synthesis can be stimulated via two pathways: (1) upon surface sensing, whereby holdfast is produced within seconds of surface contact (Li et al., 2012, Hoffman et al., 2015) or (2) developmentally, where newborn swarmer cells produce holdfast after they have completed approximately 30% of the cell cycle (Levi & Jenal, 2006). Therefore, we investigated both pathways to determine if defects in holdfast synthesis could explain the biofilm formation defects observed in the Mot- and Fla- mutants.
It has been previously proposed that increased load on the flagellum filament, caused by the simultaneous binding of the pili and rotating flagellum to surfaces, triggers the rapid synthesis of holdfast (Li et al., 2012). To determine whether the absence of a flagellum in the Fla- mutant could act on holdfast production by mimicking surface-contact stimulation, we measured the timing of holdfast synthesis after surface contact for Fla- and Mot- mutants. While previous studies used cell synchronization methods to measure holdfast synthesis after surface contact, Fla- and Mot- mutants cannot be synchronized (Li et al., 2012). To bypass this issue, we grew mixed cultures in PDMS chambers and tracked single cells as they reached the surface (Ellison et al., 2017). We observed holdfast production by microscopy by including fluorescently labeled Wheat-Germ Agglutinin (AF488-WGA) in the chambers. AF488-WGA binds specifically to the N-acetyl glucosamine residues present in holdfasts, enabling the microscopic observation of holdfast production in real-time (Merker & Smit, 1988). We recorded the difference between the time it took for a cell to reach the surface (t0) and to synthesize a holdfast (tH) (Fig. 2A). We found that in M2G defined medium, surface contact does not stimulate holdfast synthesis in WT cells (Fig 2B, top panels). Therefore, defects in surface sensing are unlikely to explain the differences in biofilm phenotype between WT and Fla- cells in this type of medium.
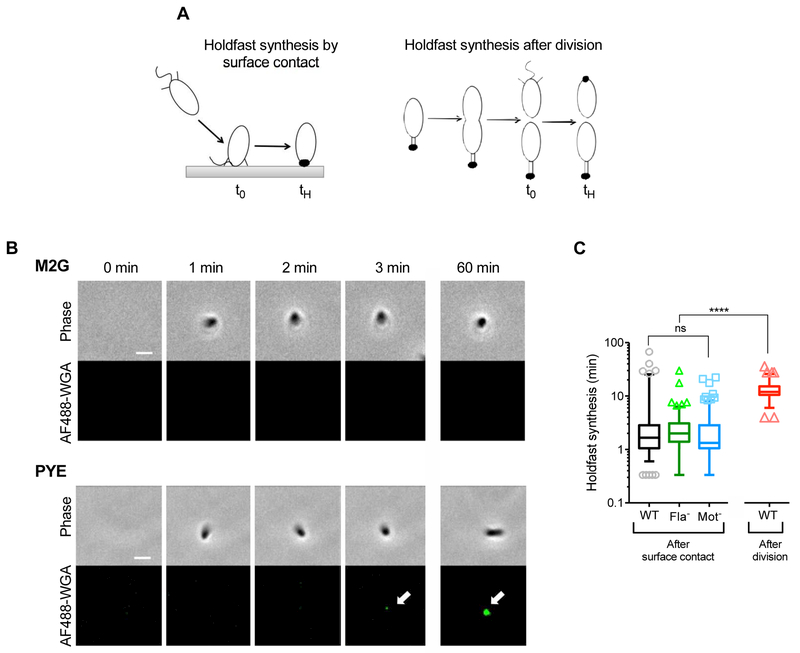
Fig. 2: Timing of holdfast synthesis after surface contact in WT and flagellar mutant populations.
(A) Schematic representation of timing of holdfast production by surface-contact stimulation or by developmental program. t0 = time when the cell first encounters the surface, in the case of surface stimulated holdfast production or time of cell division, in the case of developmentally regulated holdfast production; tH = time when holdfast is first detected on the cell by fluorescence microscopy. Holdfast synthesis timing is calculated from tH – t0. (B) Representative time-lapse images of a single NA1000 hfsA+ WT cell reaching the surface (phase channel) and producing a holdfast, visualized using AF488-WGA binding (fluorescence channel). (C) Timing of holdfast synthesis after surface contact on a glass surface in liquid compared to holdfast synthesis after division. Experimental data points for NA1000 hfsA+ WT and Mot- strains were extracted from (Ellison et al., 2017). Data are represented by box plots with 5–95% percentile whiskers, from 2 independent replicates. Statistical comparisons are calculated using Mann-Whitney unpaired t-tests. **** P < 0.0001; ns = not significant. The number of cells plotted (cells which arrived on the surface without a holdfast and produced one within 3 h) is n = 115, 156 and 182 for WT, ∆flgE and ∆motB respectively. The total number of cells tracked (including cells arriving with holdfast already synthesized) is n = 241, 257 and 566 for WT, ∆flgE and ∆motB respectively.
In PYE medium, upon surface contact, WT cells produce holdfast within approximately 1 min (Fig. 2B, bottom panels, Fig. 2C and Table 1). In comparison, cells grown in non-motile conditions on agarose pads produce holdfast approximately 12 min after division (Fig. 2C), as discussed later. Thus, surface contact stimulation occurs an order of magnitude faster than developmentally triggered holdfast synthesis (Fig. 2C and Table 1). Interestingly, not all WT cells produce holdfast rapidly after surface contact; in around 10% of cells, holdfasts are not produced until at least 10 minutes after reaching the surface (Fig. 2C), a timing corresponding to developmental regulation of holdfast synthesis (Fig. 2C, red box). Fla- and Mot- mutants behave similarly to WT upon surface contact, producing holdfast approximately 1 min after reaching the surface (Fig. 2C and Table 1). These results indicate that, under our experimental conditions, cells can undergo rapid surface contact stimulation of holdfast synthesis independent of the presence of a flagellum or its rotation.
TABLE 1:
Timing of holdfast synthesis under the different conditions used in this study.
| Holdfast synthesis after division on agarose pads (min)1 | Holdfast synthesis after surface contact in static conditions (min) 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | M2G | PYE | PYE | ||||||
| Median | SEM | n | Median | SEM | n | Median | SEM | n | |
| WT | 27 | 1.11 | 123 | 10 | 0.56 | 159 | 1.67 | 0.83 | 115 |
| Fla− | 12 | 0.72 | 116 | 8 | 0.62 | 127 | 2 | 0.24 | 156 |
| Mot− | 26 | 1.29 | 71 | 10 | 0.63 | 153 | 1.33 | 0.24 | 182 |
| Pil− | 28 | 1.48 | 75 | 24 | 1.38 | 99 | ND | ND | ND |
| Pil− Fla− | 14 | 0.86 | 83 | 22 | 1.49 | 102 | ND | ND | ND |
| HfiA− | 12 | 1.33 | 74 | 8 | 0.91 | 53 | ND | ND | ND |
| HfiA− Fla− | 8 | 0.87 | 97 | 10 | 0.93 | 46 | ND | ND | ND |
Data extracted from time-lapse microscopy of cells on agarose pads
Data extracted from time-lapse microscopy of cells reaching the glass surface on PDMS flow chambers
ND: Not determined
Because Fla- mutants synthesize holdfast upon surface contact in PYE, we hypothesized that holdfast synthesis would be independent of flagellar arrest, in contrast to previous models (Li et al., 2012). We therefore sought to analyze the timing of holdfast synthesis in cells with arrested flagellar rotation. To visualize flagella and their rotation, we constructed strains with a cysteine substitution in the major flagellin subunit FljK (Faulds-Pain et al., 2011). This enabled visualization of the flagellum by labeling with a cysteine-reactive maleimide dye, as has been previously described (Blair et al., 2008). Using a FljK structure generated by the Protein Fold Recognition Server PHYRE (Kelley & Sternberg, 2009), we predicted positions where cysteine substitutions should be exposed and not impact flagellum synthesis (threonines at positions 103 and 176) (Fig. 3A). We confirmed experimentally that the mutations FljKT103C and FljKT176C did not impair motility in low percentage agar (Fig 3B). Flagellum filaments were labeled using AF488 conjugated maleimide dye (AF488-mal) and visualized with live fluorescence microscopy, and flagellum labeling did not impede swimming in liquid cultures (Fig. 3C, Movie S1). During flagellum labeling experiments with AF488-mal, we noticed that holdfast was also labeled. Holdfast labeling by AF488-mal was specific to holdfasts attached to cells, as shed holdfasts from an hfaB holdfast anchor mutant were not labeled (Fig S2). It has been shown recently that the amine-reactive succinimidyl ester dye shows the same labeling profile (Hernando-Pérez et al., 2018). Shed holdfasts from a ∆hfaB mutant differ from holdfasts anchored to WT cells in that they lack the cysteine-containing outer membrane holdfast anchor protein HfaD (Hardy et al., 2010). The presence of cysteine-containing HfaD in cell-anchored holdfasts may explain the labeling of those holdfasts with AF488-mal.
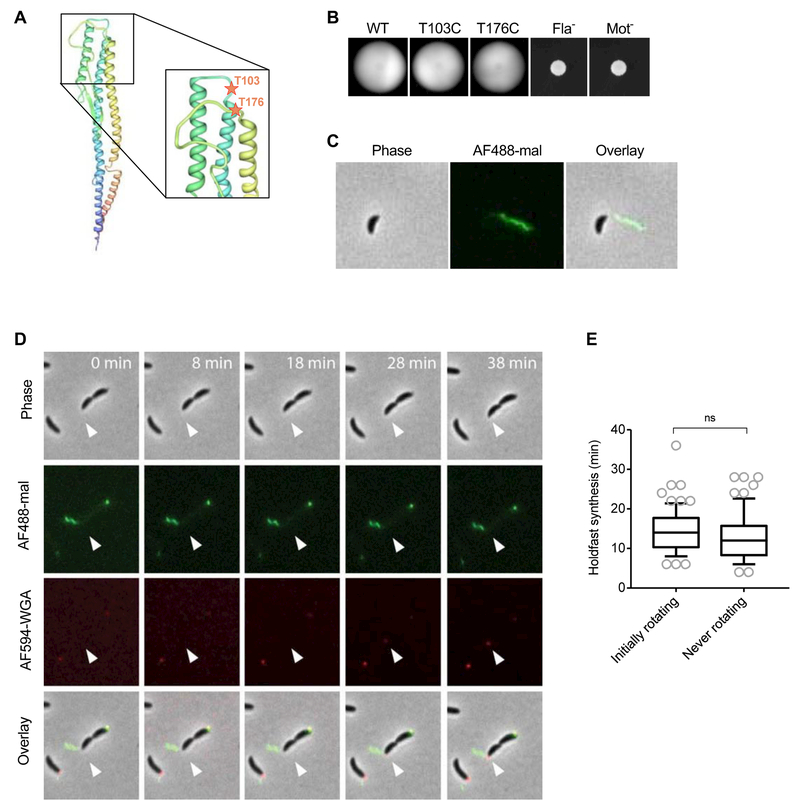
Fig. 3: Holdfast synthesis timing upon arrest of flagellar rotation.
(A) Construction of FljK-cys mutants. PHYRE predicted structure of the FljK flagellin. The N-terminus of the predicted FljK structure is blue and the C terminus is red. The positions of the mutated amino acids T103 and T176 are depicted as red stars. (B) Motility assay in low percentage PYE (0.3%) agar. (C) Staining of a swarmer cell of NA1000 hfsA+ fljKT103C using AF488-mal under a PYE 0.8% agarose pad. (D-E) Timing of holdfast synthesis by newly divided swarmer cells grown in PYE and constrained by a 0.8% agarose pad containing AF594-WGA. (D) Representative time lapse of swarmer cell with an arrested flagellum and subsequent holdfast synthesis. Cells were imaged using phase contrast microscopy. Flagella and holdfasts were stained using AF488-mal and AF594-WGA respectively. The white arrow indicates the location of expected holdfast synthesis. (E) Timing of holdfast synthesis by newly divided swarmer cells. Cells labeled as “initially rotating” are newly divided cells that exhibited a rotating flagellum that became quickly arrested under the pad; and cells labeled as “never rotating” are newly divided cells in which the flagellum did not rotate at any time. Data are represented by box plots with 5–95% percentile whiskers. n = 72 and 66 cells for rotating and non-rotating flagella respectively. No statistically significant differences between the 2 conditions can be calculated using Mann-Whitney unpaired t-tests (ns).
We analyzed the behavior of the flagellum in live cells under PYE agarose pads and monitored flagellum rotation and holdfast synthesis simultaneously. Upon imaging the FljKT103C strain on PYE agarose pads, we found that flagellar rotation was arrested in most cells. To determine whether arrest of flagellum rotation was associated with holdfast synthesis, we tracked holdfast synthesis in newly divided cells in which flagellum rotation was clearly halted (Fig. 3D and Movie S2). We analyzed newly divided cells that exhibited either a rotating flagellum that became quickly arrested under the pad or a flagellum that did not rotate at any time during the experiment. We measured the timing of holdfast synthesis after division in both populations and found no significant difference, demonstrating that arrest of flagellum rotation is not sufficient to stimulate surface sensing (Fig 3E). In summary, these results show that, under our experimental conditions, neither the flagellum nor its stator is required for surface stimulated holdfast synthesis in PYE, and that arrest of flagellum rotation does not stimulate holdfast synthesis.
Fla- mutants produce holdfast prematurely during the cell cycle in M2G.
As shown above, cells do not undergo surface contact stimulation of holdfast synthesis in M2G. We thus hypothesized that the adhesion differences between the Fla- and Mot- mutants in M2G are due to effects on developmental regulation of holdfast synthesis. To assess cell cycle progression and the timing of holdfast synthesis in Fla- mutants, we first tracked single cells and monitored cell differentiation and holdfast synthesis by time-lapse microscopy using both PYE and M2G media on agarose pads (Fig 4A and B). Previous data suggest that agarose pads still marginally stimulate holdfast production via the surface sensing response: cells lacking pili, appendages that are required for surface sensing (Ellison et al., 2017), exhibit delayed holdfast synthesis compared to WT cells when grown on PYE agarose pads (Hoffman et al., 2015). Therefore, as a control, we first tracked the timing of holdfast synthesis on agarose pads of a strain lacking pili (∆pilA, Pil-) in both media conditions (Fig. 4A and B, Table 1). While holdfast synthesis was delayed in a Pil- mutant compared to WT in PYE, this mutant exhibited the same timing of holdfast synthesis as WT in M2G, consistent with our earlier observations that surface stimulation does not occur in defined nutrient conditions. Thus, timing of holdfast synthesis on M2G agarose pads reflects developmental stimulation.
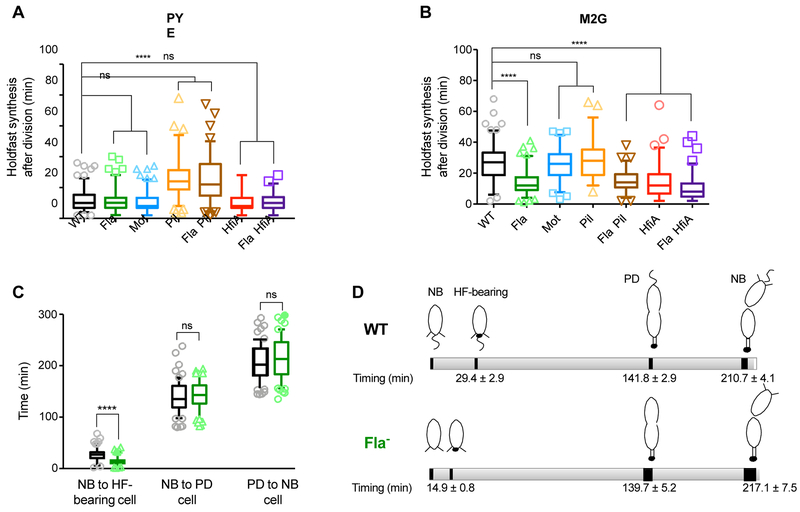
Fig. 4: Holdfast synthesis timing.
Timing of holdfast synthesis by newly divided swarmer cells on PYE (A) or M2G (B) agarose pads containing AF488-WGA. The timing between frames when a new cell divides and when the holdfast first appears on that same cell is recorded and the data are represented by box plots with 5–95% percentile whiskers. Number of cells counted is indicated in Table 1. (C) Timing of cell differentiation on M2G agarose pads. The timing between, (1) when a newborn cell (NB cell) is created upon division and when the holdfast is visualized on that same cell (HF-bearing cell); (2) when the same newborn cell differentiates into a predivisional (PD) cell; and (3) when this predivisional cell divides to form a new newborn cell, is recorded and the data are represented by box plots with 5–95% percentile whiskers. 55 and 63 cells were counted for CB15 WT (black boxes) and ∆flgE (green boxes) respectively. Statistical comparisons are calculated using Mann-Whitney unpaired t-tests. **** P < 0.0001; ns = not significant. (D) Schematic representation of cell differentiation timing on M2G agarose pads of CB15 WT (top) and ∆flgE (bottom), with timing extracted from data from Fig. 4B and4C.
On PYE agarose pads, the timing of holdfast synthesis after division for Fla- and Mot- mutants was indistinguishable from WT (Fig. 4A and Table 1). In pads containing M2G medium, WT and Mot- cells produced holdfast at roughly the same time during the cell cycle, with a mean of 27 and 26 min after cell division, respectively (Table 1, Fig. 4B). However, holdfast synthesis occurred 15 min earlier in Fla- than in WT cells in M2G, at a mean of 12 min after cell division (Table 1, Fig. 4B). We measured the timing of differentiation from newborn swarmer cells to predivisional cells, and from predivisional cells to division in WT and Fla- strains (Fig. 4C) and show that Fla- mutants and WT cells were not significantly different with respect to growth rate or timing of cell division (Fig. 4D). We conclude that, in defined medium, the Fla- mutation hastens holdfast synthesis during the cell cycle and we infer that the resulting increase in the proportion of cells bearing a holdfast underlies the adhesion phenotype of Fla- mutants. An alternate hypothesis is that WT cells are delayed for holdfast synthesis, when grown in defined medium, compared to cells grown in PYE medium, while Fla- mutants exhibit no delay in holdfast synthesis during the cell cycle due to differences in nutrient availability. Based on these results, we infer that the resulting increase in the proportion of cells bearing a holdfast underlies the adhesion phenotype of Fla- mutants. The premature holdfast synthesis observed in the Fla- mutant, but not the Mot- strain, clarifies the difference in biofilm phenotypes observed between these mutants in defined M2G medium.
It is interesting to note the behavior of a Fla- Pil- double mutant strain under different nutrient conditions. In PYE, where pilus-dependent surface contact can stimulate holdfast synthesis, the double Fla- Pil- mutant phenocopies the Pil- mutant. However, in M2G, where only developmentally regulated holdfast synthesis is observed, the double mutant phenocopies a Fla- mutant (Fig. 4A and B, Table 1). We conclude that (1) the pili, and not the flagellum, are the main determinant of surface stimulated holdfast synthesis in static conditions, and (2) premature holdfast elaboration in strains lacking a flagellum is due to aberrant developmental regulation.
Fla- mutant populations produce more holdfasts in M2G in planktonic cultures.
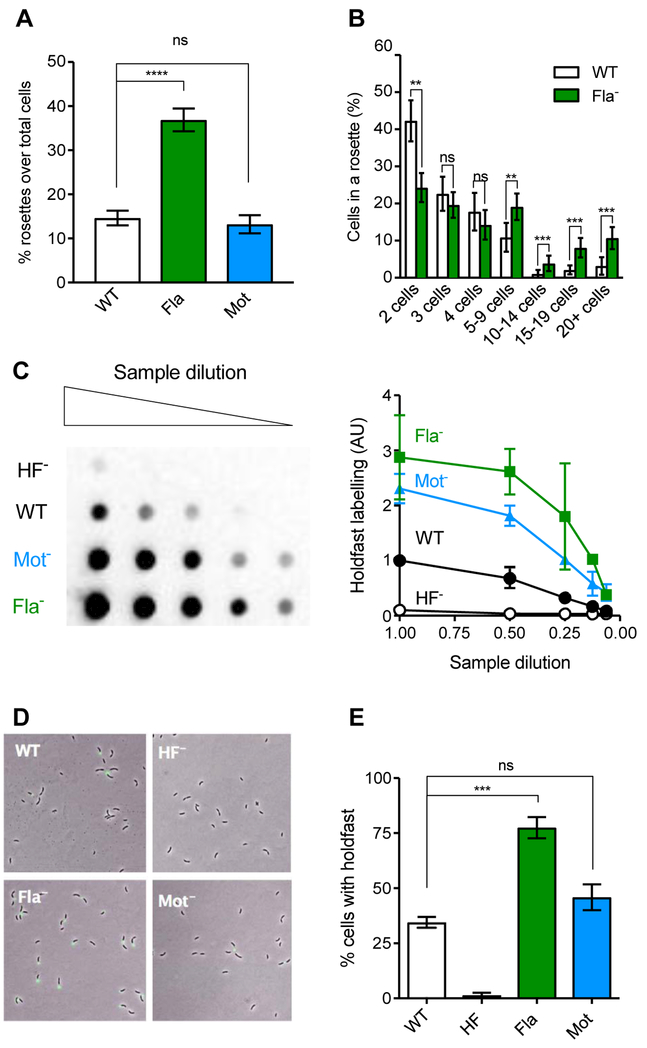
To further test if holdfast overproduction by the Fla- mutant in M2G is linked to developmental regulation, we analyzed holdfasts in planktonic cells, in the absence of surface stimulation. Holdfast production can be indirectly quantified in planktonic cultures by observing the occurrence of rosettes, multicellular complexes of cells attached to one another by their holdfasts. Quantification of rosette numbers in mid-exponential populations showed that Fla- populations formed significantly more rosettes than the WT and the Mot- cultures (Fig. 5A). Moreover, rosettes of the Fla- mutant were larger in size than those in WT cultures (Fig. 5B). These results indicate an enhancement of holdfast-mediated cell-cell interactions in a Fla- population in M2G and suggest that this mutant produces more holdfast than WT at the population level. To directly quantify overall holdfast polysaccharide production, total holdfast material was extracted using cell extracts from mid-log phase (OD600 of 0.4 – 0.8) cultures grown in M2G and spotted onto nitrocellulose membranes. The dot-blots containing holdfast extracts were probed with HRP-conjugated WGA lectin. Lectin blot and pixel intensity quantification (Fig. 5C) revealed that the Fla- mutant produces approximately three times more holdfast polysaccharide than WT at the population level. Mot- holdfast polysaccharide levels were also elevated, although not to the same extent as the Fla- mutant.
Fig. 5: Holdfast production in mixed populations using M2G medium.
(A) Average percentage of cells in rosettes formed by cells in culture; n = 780, 463 and 650 for CB15 WT, ∆motB (Mot-), and ∆flgE (Fla-) respectively. (B) Number of cells in each rosette for WT (white) and ∆flgE (green). The error is represented as SEM. Statistical comparisons are calculated using Mann-Whitney unpaired t-tests. **** P < 0.0001; *** P < 0.001; ** P < 0.01; ns = not significant. (C) Left, representative lectin blot of total holdfast produced by Fla- and Mot- mutants (Columns are two-fold dilution series), using HRP-WGA binding to visualize holdfasts by chemiluminescence. Right, pixel intensity quantification of 3 different blots normalized to the signal intensity of the WT sample at the highest concentration, with error represented as SEM. C. crescentus WT (solid black circles), ∆motB (Mot-, blue solid triangles), CB15 ∆flgE (green solid squares) and ∆hfsDAB (HF-, black open circles). (D) Representative images of cells grown to mid-exponential phase in PYE and stained with AF488-WGA to visualize holdfasts. (E). Quantification of cells harboring a holdfast in mixed populations. The results represent the average of at least four independent replicates (more than 500 cells per replicate) and the error bars represent the SEM. Statistical comparisons are calculated using Student’s unpaired t-tests. *** P < 0.001; ns = not significant.
To determine if the Fla- mutation increases the proportion of cells harboring a holdfast, we stained early-exponential cultures grown in M2G with fluorescent WGA and quantified the number of cells with a holdfast in the mixed population by fluorescence microscopy (Fig 5D). Approximately 35% of WT cells possessed a holdfast (Fig. 5E). The proportion of cells bearing holdfast was not significantly increased in the Mot- mutant (Fig. 5E). However, in Fla- cultures, 75% of cells exhibit a holdfast (Fig 5E). Thus, in defined growth medium, Fla- mutant cells develop holdfast earlier in the cell cycle, resulting in a greater portion of cells harboring holdfast at a population level.
The nutrient-dependent premature holdfast synthesis by Fla- mutant is c-di-GMP-dependent and is regulated by the HfiA pathway.
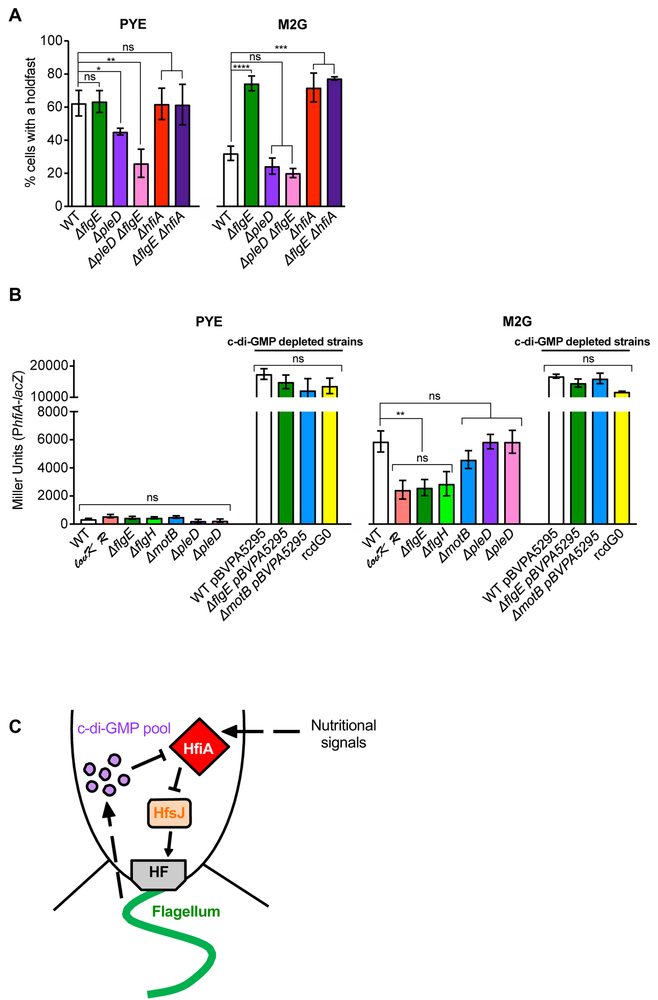
c-di-GMP is a second-messenger well-known to regulate both holdfast synthesis and flagellum assembly (Jenal & Malone, 2006). The diguanylate cyclase response regulator PleD is involved in biofilm formation and proper timing of holdfast synthesis during the cell cycle in PYE (Levi & Jenal, 2006, Abel et al., 2011, Sprecher et al., 2017) and is required for flagellum ejection (Aldridge et al., 2003). Deletion of the pleD gene reduces cellular c-di-GMP concentrations by approximately a third (Abel et al., 2013), making it an ideal candidate for testing the effects of reduced c-di-GMP on the holdfast phenotypes observed in the Fla- mutant. We hypothesized that misregulation of holdfast production timing in Fla- mutants might be a result of misregulation of c-di-GMP levels through pleD. Therefore, we investigated the behavior of a ∆pleD and a ∆pleD ∆flgE double mutant for holdfast production in both PYE and M2G (Fig. 6A). In PYE, a ∆pleD population produced fewer holdfasts than WT (Fig. 6A), as previously reported (Abel et al., 2011). Compared to PYE, the number of cells harboring a holdfast in M2G decreased by a factor of about two in both WT and ∆pleD populations: 62% versus 32% for WT and 45% versus 24% for ∆pleD populations respectively (Fig. 6A). We interpret that the response to changes in nutrient availability is not altered in a ∆pleD mutant. In M2G, the ∆pleD ∆flgE double mutant did not display the hyper-holdfast phenotype of the ∆flgE single mutant, but rather phenocopied the slightly hypo-holdfast phenotype of the ∆pleD population (Fig. 6A). The number of holdfasts produced by the single ∆flgE and ∆pleD mutants could be restored to WT levels, when a copy of flgE and pleD respectively was expressed in trans by a replicating plasmid (Fig. S3A). We infer that c-di-GMP is involved in the misregulation of holdfast synthesis in the Fla- mutants. Interestingly, we noticed increased variation in the amount of holdfast produced in the ∆pleD ∆flgE strain containing the empty plasmids or complementing vectors, grown in the presence of kanamycin (Fig. S3A), compared to the same strain without a plasmid (Fig. 6). This result is puzzling and illustrates the complexity of this regulation. However, complementation of the ∆pleD ∆flgE double mutant with an ectopic copy of pleD restored holdfast synthesis to ∆flgE mutant levels, supporting earlier observations that increased holdfast synthesis in the Fla- mutant is dependent upon PleD.
Fig. 6: Role on HfiA in Fla- holdfast misregulation.
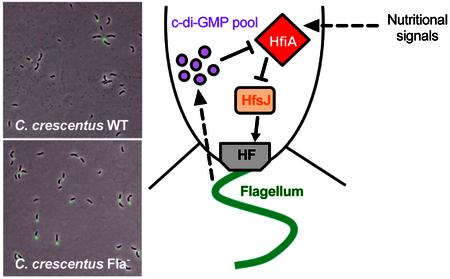
(A) Quantification of cells harboring a holdfast in mixed populations grown to exponential phase. The results represent the average of at least two independent replicates (more than 250 cells per replicate) and the error bars represent the SEM. Statistical comparisons are calculated using Student’s unpaired t-tests. ** P < 0.01; * P < 0.1; ns = not significant. (B) ß-galactosidase activity of PhfiA-lacZ transcriptional fusions in PYE (left) and M2G (right). The results represent the average of at least 6 independent cultures (assayed on at least 2 different days) and the error bars represent the SEM. Statistical comparisons are calculated using Student’s unpaired t-tests. ** P < 0.01; * P < 0.1; ns = not significant. (C) Proposed model. Defects in flagellum synthesis result in an increase of c-di-GMP, which reduces transcription of the holdfast synthesis inhibitor gene hfiA, thereby resulting in an increase in holdfast synthesis.
To determine if c-di-GMP levels inside the cell can impact the response observed in the Fla- mutant, we sought to modulate c-di-GMP levels by transforming cells with a replicating plasmid bearing the gene encoding the phosphodiesterase (PDE) PA5295 from Pseudomonas aeruginosa under the control of the vanillate inducible promoter (pBV-PA5295) (Duerig et al., 2009). It has been reported previously that expression of the PDE PA5295 in C. crescentus depletes the intracellular levels of c-di-GMP below detection when the promoter is induced by 50 mM vanillate (Duerig et al., 2009), comparable to a mutant where all the known encoding diguanylate cyclases and PDEs are deleted, the rcdG0 strain (Hug et al., 2017). We measured the amount of biofilm formed by the strains overexpressing the PA5295 PDE and found that WT, Fla-, and Mot- strains are unable to form a biofilm in either PYE or M2G, similarly to the c-di-GMP null strain rcdG0 (Fig. S3C). These results suggest that the increased attachment observed in Fla- mutants may be due to an increased production of intracellular c-di-GMP. To determine if Fla- mutants produce more c-di-GMP, we quantified the amount of intracellular c-di-GMP levels in the different strains (Fig. S3C). As previously reported, the ∆pleD mutant exhibited a reduction of c-di-GMP by a third compared to WT cells (Abel et al., 2013). Interestingly, there were no signifcant differences in c-di-GMP levels detected between populations of Fla-, Mot-, or WT strains in either PYE or M2G media. We speculate that changes in c-di-GMP levels responsible for observed changes in biofilm and holdfast phenotypes may be masked by heterogenous populations of cells that contain cells in all stages of the cell cycle. It is also possible that a local pool of c-di-GMP is important for this regulation, and not the overall c-di-GMP concentration in the cell.
The transcription of the holdfast regulatory gene hfiA is reduced in a PleD-dependent manner in Fla- mutant in defined medium.
In C. crescentus, holdfast synthesis is inhibited in M2G defined medium by elevated transcription of the gene encoding the holdfast inhibitor protein HfiA (Fiebig et al., 2014). Deletion of hfiA derepresses holdfast synthesis in defined nutrient conditions. Because of the similar phenotype observed in the Fla- mutant, and because of the stark contrast in the timing of holdfast synthesis of the Fla- mutant in defined vs complex medium, we investigated whether hfiA is involved in the increase in holdfast synthesis in the Fla- mutant. We first quantified the number of cells harboring a holdfast in a ∆hfiA mutant (HfiA-) or a HfiA- Fla- mutant mixed populations in both complex PYE and defined M2G media (Fig. 6A). The ∆flgE and ∆hfiA single mutants and the ∆hfiA ∆flgE double mutant were indistinguishable, with around 60% and 75% of cells harboring a holdfast in PYE and M2G respectively, compared to around 50% and 30% for WT cells in those media, respectively (Fig. 6A). We also measured the timing of holdfast production in newborn cells grown on PYE and M2G agarose pads. Again, the ∆hfiA and ∆hfiA ∆flgE strains phenocopied the Fla- mutant: they produced holdfast prematurely in M2G, but not in PYE (Fig. 4 and Table 1). Since the single mutant phenotypes are similar, and the ∆hfiA ∆flgE double mutant phenotype is not additive, we infer that these genes are in the same pathway.
We hypothesized that the Fla- mutations affected holdfast synthesis by misregulation of hfiA. If flagellum mutations reduce hfiA expression, holdfast synthesis would be depressed. To test the validity of this model, we monitored the expression of hfiA using lacZ transcriptional fusions in WT, Fla- and Mot- strains. As a control, we measured the expression of hfiA in a lovK-lovR overexpressing strain (CB15 xyl::pMT585-lovR vanR::pMT528-lovK / pRKlac290-PhfiA; lovK+R+ PhfiA-lacZ), as a decrease in hfiA promoter activity has been previously reported in this strain in defined medium conditions (Fiebig et al., 2014). In PYE, where complex biomolecules are readily available, the activity of the hfiA promoter is minimal, and no significant difference is detected between WT and mutant strains (Fig. 6B). In WT cells, we observed an increase in hfiA transcription in M2G defined medium compared to PYE (Fig. 6B); a more modest increase occurs in the hyper-adhesive lovK-lovR overexpressing strain, in agreement with previous studies (Fiebig et al., 2014). Similar to the lovK-lovR overexpressing strain, hfiA transcription was reduced in the ∆flgE mutant compared to WT in M2G (Fig 6B). The same result was obtained using a ∆flgH mutant, another Fla- strain which exhibits a similar increased biofilm behavior (Fig. 1D). These results are consistent with a model in which flagellar mutations feedback to repress hfiA expression and thereby derepress holdfast synthesis in defined growth medium.
The ∆pleD mutation, which abrogated the enhanced holdfast phenotype in Fla- strains (Fig. 6A) also prevented the reduction of hfiA transcription by the Fla- mutation (Fig. 6B), suggesting that c-di-GMP is required for the flagellum synthesis feedback on holdfast synthesis. To test whether a reduction of c-di-GMP would affect hfiA transcription, we overexpressed the PA5295 PDE as described above. We found that in both PYE and M2G, these strains exhibited a very high level of transcription of hfiA including in the c-di-GMP null rcdG0 strain used as a control (Fig. 6B). These results suggest that repression of hfiA is c-di-GMP dependent.
Taken together, these results are consistent with a model in which a fully assembled flagellum promotes synthesis of the holdfast inhibitor, HfiA, and thereby represses holdfast synthesis and surface attachment. In this model, a partially assembled or disassembled flagellum feeds back to repress hfiA expression through a mechanism that requires c-di-GMP. One prediction of this model is that ectopic overexpression of hfiA should overcome holdfast and adhesion phenotypes of the flagellar mutants. To test this, we chromosomally inserted hfiA at the xylX locus in the Fla- and Mot- strains and measured biofilm formation in strains grown in PYE or M2G that were induced for hfiA expression (Fig. S4). Indeed, induction of hfiA reduces biofilm production in all strains regardless of nutrient conditions, by 75 to 90% of WT empty vector strain. It is worth noticing that, in M2G, the impact of hfiA overexpression is statistically less important for the Fla- mutant than for the WT and Mot- strains (Fig. S4). These results further support the model that hfiA is downstream of the flagellum feedback regulation of holdfast synthesis (Fig. 6C).
DISCUSSION
In this work, we investigated the role of the flagellum in the switch between motility and sessility in C. crescentus. We showed that Fla- mutants display enhanced biofilm formation due to premature holdfast synthesis during the cell cycle in defined medium and that this phenotype is independent of surface-stimulated holdfast synthesis.
Many studies have suggested that obstruction of flagellum rotation triggers surface-associated phenotypes such as adherence and secretion of exopolysaccharides by acting as a surface-contact mechanosensor (Belas, 2014). However, our results indicate that in neither the flagellum itself nor its rotation is required for surface-induced stimulation of holdfast synthesis. Instead, surface contact-triggered holdfast elaboration can occur independently of flagellum rotation or the presence of a flagellum filament (Ellison et al., 2017, Hug et al., 2017). Furthermore, surface-induced stimulation does not occur in minimal growth conditions. These results are at odds with prior hypotheses that C. crescentus surface sensing is due to an increased load on the flagellum caused by the simultaneous binding of the flagellum and pili to the surface (Li et al., 2012). Indeed, increasing viscosity through the addition of polyethylene glycol or Ficol stimulated holdfast synthesis in planktonic cultures, consistent with that model (Li et al., 2012). How can these results be reconciled? It was subsequently shown that in C. crescentus, an increase in viscosity impacts cell viability as evidenced by their membrane leakage (Gao et al., 2014). Membrane perturbation stimulates surface-associated behaviors in V. cholerae (Van Dellen et al., 2008), suggesting that membrane potential plays a role in surface sensing. The effect of increased viscosity on membranes rather than on the flagellum may stimulate surface sensing in C. crescentus. Indeed, a recent study indicates that the flagellum motor, but not its filament, is important for mediating surface attachment in C. crescentus under constant flow (Hug et al., 2017), raising the possibility that the Mot complex could serve as a mechanosensor independent of flagellum rotation. While our results show that neither the flagellum nor its motor is essential for holdfast synthesis upon surface contact in static conditions, the increased shear force experienced by surface-associated cells under flow may stimulate holdfast synthesis by a membrane-perturbation pathway dependent on the flagellum stator.
It has previously been observed that under agarose pads, Pil- mutants display delays in holdfast synthesis (Hoffman et al., 2015), a result that we confirm here. Coupled with recent findings that pili are required for surface sensing and that resistance on pilus retraction is used to sense surfaces (Ellison et al., 2017), these data suggest that WT cells are capable of a low level of surface-stimulation of holdfast synthesis under PYE pads. Additionally, the Pil- strain exhibits the same timing of holdfast synthesis in defined M2G medium as WT cells, an observation that is consistent with our finding that cells do not make holdfast in response to surface contact under those conditions. The timing of holdfast synthesis on agarose pads in a Fla- Pil- mutant phenocopies the behavior of the Fla- single mutant in defined medium, further implicating an aberrant developmental regulation of holdfast synthesis in Fla- strains in defined medium.
An interesting result of our study is that the long-term adhesion phenotypes of Fla- and Mot- mutants are different in defined nutrient conditions. While Fla- mutants form as much biofilm as WT over time, Mot- mutants are strongly deficient at every stage of biofilm formation. Similar observations have been made in other microorganisms including Yersinia enterocolitica, Bacillus cereus, Agrobacterium tumefaciens and Listeria monocytogenes, where Fla- mutants adhere more efficiently to surfaces than Mot- strains (Merritt et al., 2007, Kim et al., 2008, Houry et al., 2010, Todhanakasem & Young, 2008). Our data suggests that the increased attachment of Fla- mutants is due to increased holdfast synthesis promoting adhesion before dispersion can occur. In contrast, in Mot- mutants, do not exhibit as much of an increase in holdfast synthesis and furthermore, the presence of a bulky paralyzed flagellum filament may be impairing holdfast / surface interaction. This may explain the consistently impaired adhesion phenotype of Mot- mutants in contrast to Fla- mutants.
It is well established that cell adhesion and biofilm formation and architecture can be influenced by nutrient availability (Flemming & Wingender, 2010). For example, nutrient limitation (phosphate-limiting conditions) enhances the production of the unipolar polysaccharide UPP in A. tumefaciens (Xu et al., 2012) and Rhodopseudomonas palustris (Fritts et al., 2017). In this work, we show that increased holdfast synthesis in a Fla- mutant is dependent on environmental conditions and nutritional environment: the misregulation of holdfast synthesis in Fla- mutants correlates with misregulation of HfiA, which is mediated through PleD, a known diguanylate cyclase response regulator that likely mediates its effects through modulation of c-di-GMP. Interestingly, there are no obvious effects of flagellar mutations on holdfast synthesis in PYE medium, suggesting that this alteration of c-di-GMP is also medium-specific. Although measurements of c-di-GMP revealed no significant differences between either media or between Fla- and WT strains, our results suggest that c-di-GMP changes are involved in the misregulation of holdfast synthesis under nutrient defined conditions. These data could suggest two things: (1) that subtle changes in either intracellular concentrations or subcellular localization of c-di-GMP are sufficient to mediate drastic changes in heterogenous populations of cells, or (2) that another player is involved in the regulation of hfiA transcription in a c-di-GMP dependent mechanism.
Thus far, HfiA is the only known regulator of holdfast synthesis that responds to nutrient environment. The mechanism by which nutrient status regulates hfiA transcription is unknown. It will be interesting to determine if flagellar mis-assembly and nutrient status signals converge upstream of hfiA regulation or if these signals independently tune HfiA expression. Interestingly, we found that the increase in holdfasts and rosettes in Fla- mutants is greater than would be expected based on observed reduction of hfiA transcription alone. It is therefore possible that the absence of the flagellum may be playing an additional role in post-transcriptional regulation of HfiA or the holdfast machinery itself.
Flagellum biosynthesis is tightly controlled by a cell-cycle regulated hierarchal program in which late flagellar genes are not transcribed until early components of the flagellum apparatus are produced (Wu & Newton, 1997, Brown et al., 2009). This biosynthesis pathway is regulated by the σ54 transcriptional activator FlbD and its cognate repressor FliX that prevents gene transcription in the absence of early flagellar machinery production (Muir & Gober, 2004). Additional flagellum regulatory components include FlbT, a post-transcriptional regulator that controls the production of flagellins by monitoring the status of flagellum assembly (Anderson & Gober, 2000). These regulatory systems allow for strict control of the production of flagellum components and may provide a mechanism by which flagellum synthesis can regulate other important processes including the timing of holdfast synthesis that we report here. Other σ54-dependent motility master regulators, FlrA in Vibrio cholerae or FleQ in P. aeruginosa, have been shown to bind to c-di-GMP to regulate the expression of flagellar genes, or genes involved in polysaccharide production respectively (Srivastava et al., 2013, Hickman & Harwood, 2008). A similar mechanism could occur in C. crescentus, where c-di-GMP binding to one or several master regulators could control of flagellum biosynthesis and holdfast production in concert. In addition, it may be advantageous to couple flagellum biosynthesis and holdfast production under some environmental conditions. For example, surfaces accumulate nutrients (Costerton et al., 1995), an especially important consideration for freshwater organisms in which the bulk fluid is nutritionally poor. In the absence of motility, which is often used to guide cells towards nutrient sources, surface attachment may improve survival under nutrient-limited conditions.
In conclusion, we have demonstrated that, in C. crescentus, there is feedback regulation between flagellum synthesis and holdfast production. Mutants lacking flagella adhere more efficiently to surfaces, as they synthesize holdfast earlier during the cell cycle, dependent on nutrient availability and independent of surface contact. We show that feedback regulation of holdfast synthesis by flagellum synthesis occurs by reducing expression of the holdfast inhibitor protein HfiA and that this reduction depends on PleD, suggesting mediation by c-di-GMP (Fig. 6C). Further exploration of this link may reveal how bacteria coordinate cell cycle processes in various nutrient conditions and how this process optimizes adhesion and survival in various environments.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and growth conditions.
The bacterial strains used in this study are listed in Table S1. C. crescentus strains were grown at 30˚C, in defined M2 medium supplemented with 0.2% (w/v) glucose (M2G) (Johnson & Ely, 1977) or Peptone Yeast Extract (PYE) medium (Poindexter, 1964). When appropriate, antibiotics were added to the following concentrations: kanamycin 5 µg/ml, chloramphenicol 1 µg/ml, gentamycin 2.5 µg/ml and tetracyclin 1 µg/ml. For induction of the xyl::pMT680-hfiA strains, 0.3% xylose was added to the culture prior to overnight incubation. 50 mM vanillate was added to the culture for induction of the pMT630, pMT630-motB, pMT630-pleD, pMT335, and PBV-PA5295 containing strains.
E.coli DH5α, DH10ß or TOP10 were used for cloning and grown in Luria Broth (LB) medium at 37˚C, with the following antibiotics: kanamycin 25 µg/ml, chloramphenicol 20 µg/ml, gentamycin 15 µg/ml and tetracyclin 10 µg/ml, when appropriate. Plasmids were transferred in C. crescentus by electroporation or by mating using E.coli S17.1 (Ely, 1991).
In-frame deletion mutants were obtained by double homologous recombination as previously described (Ried & Collmer, 1987). Briefly, genomic DNA was used as the template to PCR-amplify 500 bp fragments from upstream and downstream regions of the gene to be deleted. The primers designed for these in frame deletions are listed in Table S2. For the flgE, motB, and pilA in-frame deletions, PCR fragments were gel-purified and then digested by BamHI and XhoI or XhoI and HindIII for upstream and downstream fragments respectively. Purified digested fragments were then cloned into the suicide vector pNPTS138 and digested by BamHI and HindIII. For the flgH in-frame deletion, The up and down fragments were amplified individually, then joined by overlap extension PCR to generate a PCR product with the null allele. This PCR product was purified, digested with BamHI and EcoRI and ligated into pNPTS138. The pNPTS138-based constructs were transformed into E. coli and introduced in the host C. crescentus by mating, or directly electroporated in C. crescentus. The two-step recombination was carried out using sucrose resistance and kanamycin sensitivity. Then, the mutants were checked by sequencing to confirm the presence of the deletion. The double mutants CB15 ∆pilA ∆flgE, CB15 ∆pleD ∆flgE and CB15 ∆hfiA ∆flgE were conceived by double recombination of pNPTSflgE in the CB15 ∆pilA, CB15 ∆pleD and CB15 ∆hfiA strains respectively.
The allelic exchange constructs for the fljKT103C, fljKT107C and fljKT176C alleles were generated using double recombination as previously described (Ried & Collmer, 1987). The fljK gene was amplified from C. crescentus NA1000 hfsA+ using PCR primers fljK5’and fljK3’. The amplified region was digested with EcoRI and BamHI and ligated into similarly digested pSKII plasmid. The ligated vector was then PCR-amplified with the primers that contained the desired point mutation (Table S2). Once the point mutation sequence was confirmed (using T7 promoter and M13 reverse PCR primers, Table S2), the fljK region was extracted from the pSKII plasmid using EcoRI and BamHI, and ligated into a similarly digested pNPTS139. The resulting plasmid containing the insertions were electroporated into C. crescentus NA1000 hfsA+. The two-step recombination was carried out using sucrose resistance and kanamycin sensitivity as described above.
Motility assays.
Motility assays were performed using low percentage agar plates. Cells were stabbed in 0.3% agar PYE plates containing 50 mM vanillate and/or 5 µg/ml kanamycin when appropriate. Plates were incubated in a humid chamber at 30˚C for 5 days. The diameter of the swimming ring formed by each tested strain was measured by hand and normalized to the one of WT (with empty vector when appropriate).
Static biofilm assays.
Biofilm assays in multi-well plates were performed using two different set-ups, yielding similar results (Berne et al., 2010): (1), adhesion on polyvinyl chloride (PVC) microscope coverslips placed vertically in 12-well plates (Merritt et al., 2007); or (2), adhesion to the inside of the wells of an untreated 24-well plate (Berne et al., 2010). In both set-ups, bacteria were grown to mid-log phase (OD600 of 0.4 – 0.8) in M2G or PYE and diluted to an OD600 of 0.05 in the same medium in 3 or 0.5 ml for the 12 or 24-well plate set-up respectively, and incubated at 30˚C. For quantification, biofilms attached to coverslips or wells were rinsed with distilled H2O to remove non-attached bacteria, stained using 0.1% crystal violet (CV) and rinsed again with dH2O to remove excess CV. The CV was dissolved into 10% (v/v) acetic acid and quantified by measuring the absorbance at 600 nm (A600). The biofilm formation was normalized to A600 / OD600 and expressed as a percentage of WT.
Holdfast quantification using fluorescently labeled WGA lectin.
The number of cells harboring a holdfast was quantified by fluorescent microscopy. Holdfasts were detected with AlexaFluor 488 conjugated wheat germ agglutinin (AF488-WGA) since WGA binds specifically to the N-acetylglucosamine residues present in the holdfast (Merker & Smit, 1988). To quantify the numbers of single cells harboring a holdfast in a mixed population, early exponential phase cultures (OD600 of 0.2 – 0.4) were mixed with AF488-WGA (0.5 µg/ml final concentration), while for rosette quantification experiments, mid-exponential phase cultures (OD600 of 0.5 – 0.7) were used. In both cases, 1 µl was mounted between a slide and a coverslip sealed using valap (1:1:1 vaseline:lanolin:paraffin). Holdfasts were imaged by epifluorescence microscopy using an inverted Nikon Ti-E microscope with a Plan Apo 60X objective, a GFP/DsRed filter cube, an Andor iXon3 DU885 EM CCD camera and Nikon NIS Elements imaging software. The number of individual cells with a holdfast was calculated using the MicrobeJ plugin (Ducret et al., 2016) in ImageJ (Schneider et al., 2012). The number of rosettes and the number of cells present in a rosette were assessed manually from microscopy images.
Holdfast synthesis timing by time-lapse microscopy.
Cell division and holdfast synthesis timing were observed in live cells on agarose pads by time-lapse microscopy as described previously (Hoffman et al., 2015) with some modifications. A 1 µl aliquot of exponential-phase cells (OD600 of 0.4 – 0.8) was placed on top of a pad containing 0.8% agarose in M2G or PYE and AF488-WGA (0.5 µg/ml final) sealed under a coverslip with valap. Time-lapse microscopy images were taken every 2 min for 8–12 h using a Nikon Eclipse 90i with a 100 x DIC oil immersion objective, a Nikon 83300 filter cube T-D-F and a Photometrics Cascade K1 camera, or an inverted Nikon Ti-E microscope and a Plan Apo 60X objective, a GFP/DsRed filter cube, and an Andor iXon3 DU885 EM CCD camera. Time-lapse movies were visualized in ImageJ (Schneider et al., 2012) to manually assess the time of cell division (t0), as t = 0 when a cell newly divides, and the time of holdfast production (tH), as the first frame where the fluorescent lectin signal is detected.
Lectin blot for quantification of total holdfast produced.
Lectin blots were performed as described previously (Javens et al., 2013), with a few modifications. 1 ml of M2G-grown cells in mid-log phase (OD600 of 0.4 – 0.8) was centrifuged for 5 min at 7,500 x g. Cells were washed with 10 mM Tris HCl buffer, centrifuged for 5 min at 7,500 x g and resuspended in 100 µl of 10 mM Tris HCl buffer. After addition of proteinase K (0.5 mg/ml final concentration), the samples were incubated for 4 h at 55 °C and then at 70˚C for 20 min (proteinase K deactivation). 5 µl of benzonase (2.50 units/µl final concentration) was added to each sample and incubated at room temperature for 30 min. Serial dilutions (1:10) of samples were then made in TBS buffer pH 7.4 (10 mM Tris HCl, 150 mM NaCl) to make five dilutions per sample. 400 µl from each dilution was filtered through a nitrocellulose membrane (pre-soaked with TBS and drained) using a Biorad Bio-Dot microfiltration apparatus and rinsed with 400 µl TBS buffer. The nitrocellulose membrane was then blocked with 3% BSA in TBS 0.1% Tween 20 (TBST) for 1.5 h before adding horseradish peroxidase (HRP) conjugated-WGA lectin (5 mg/ml, 200 HRP units/ mg, Sigma) diluted to a 1:17,000 dilution and incubating for 1 h at room temperature. The blot was washed three times with TBST every 15 min. Finally, the membrane was developed using SuperSignal West Pico Substrate (Pierce, Thermo Fisher Scientific) and imaged using a Biorad ChemiDoc apparatus. Signal intensity was analyzed using ImageJ (Schneider et al., 2012).
Holdfast detection after surface contact under static conditions.
Cell cultures were grown to mid-exponential phase (OD600 = 0.4–0.6) in PYE or M2G and 200 µl of culture was diluted into 800 µl fresh medium in the presence of 0.5 µg/ml AF488-WGA for holdfast labeling. One ml of each diluted culture was flushed into a microfluidic device containing a 10 µm high chamber fabricated in PDMS (Polydimethylsiloxane) as described previously (Hoffman et al., 2015). After injection of cells into the microfluidic chamber, the flow rate was adjusted so that attachment could be observed under static conditions. Cells were grown under these conditions within PDMS devices for a 3 h period to allow for division of new cells, during which time-lapse imaging was performed. Cells that hit the surface and attached permanently via holdfast during this time period were analyzed for the timing of holdfast synthesis after surface arrival for each cell.
Time-lapse microscopy was performed using an inverted Nikon Ti-E microscope and a Plan Apo 60X objective, a GFP/DsRed filter cube, an Andor iXon3 DU885 EM CCD camera and Nikon NIS Elements imaging software. Time-lapse videos were collected for strains over a period of 3 h at 20-second intervals. Cell attachment was detected at the glass-liquid interface within the microfluidic chamber using phase contrast microscopy while holdfast synthesis was detected using fluorescence microscopy. The time difference between holdfast synthesis and cell-surface contact was determined using MicrobeJ (Ducret et al., 2016). Cells that were present on the surface at the start of the time-lapse experiment were not analyzed. 40 cells for each strain were analyzed for surface-contact stimulation of holdfast synthesis across two independent replicates.
Imaging and labeling of flagellum filaments.
A strain containing a cysteine-knock in the major flagellin fljK was used for flagellum filament staining as described previously (Blair et al., 2008) with some modifications. Cell cultures were grown to early to mid-exponential phase (OD600 = 0.2–0.4) in PYE medium. One ml of culture was mixed with maleimide conjugated to AF488 (AF488-mal) for a final dye concentration of 5 µg/ml and incubated for 5 min at room temperature. Cells were centrifuged at 8,000 x g for 1 min, washed with 1 ml of fresh PYE to remove excess dye and shed flagella, and resuspended in 50 µl of fresh PYE. One µl of the labeled culture was added to a coverslip and imaged under a 0.8% agarose PYE pad containing 0.5 µg/ml AF594-WGA to label holdfasts, and coverslips were then sealed with valap. Imaging was performed using an inverted Nikon Ti-E microscope and a Plan Apo 60X objective, a GFP/DsRed filter cube, an Andor iXon3 DU885 EM CCD camera and Nikon NIS Elements imaging software. Cell bodies were imaged using phase contrast microscopy while flagella and holdfasts were imaged using epifluorescence microscopy. Time-lapse videos were produced by imaging cells every 2 min for at least one hour.
Visualization of purified holdfasts attached on a surface.
Purified holdfasts attached to a surface were visualized as described previously (Berne et al., 2013), with few modifications. Briefly, C. crescentus CB15 ∆hfaB cells grown to late exponential phase (OD600 of 0.6 – 0.8) were pelleted by centrifugation (30 min at 4,000 x g). 100µl of supernatant, containing free holdfasts shed by the cells, were spotted on a glass coverslip and incubated for 16 h at room temperature in a saturated humidity chamber. After incubation, the slides were rinsed with dH2O to remove unbound material and labelled using AF594-WGA and AF488-mal, as described above.
β-galactosidase assays to assess hfiA promoter activity.
Strains bearing transcriptional reporter plasmids were inoculated from freshly grown colonies into 5 ml M2G or PYE medium containing 1 μg/ml tetracycline and incubated at 30˚C. Overnight cultures were diluted in the same culture medium to OD600 of 0.05 and incubated until an OD600 of 0.18 – 0.24 was reached. β-galactosidase activity was measured colorimetrically as described previously (Miller, 1972). Briefly, 200 µl of culture was mixed with 600 µl Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM ß-mercaptoethanol). Cells were then permeabilized using 50 µl chloroform and 25 µl 0.1% SDS. 200 µl of substrate o-nitrophenyl- β-D-galactoside (4 mg/ml) was added to the permeabilized cells. Upon development of a yellow color, the reaction was stopped by raising the pH to 11 with addition of 400µl of 1M Na2CO3. Absorbance at 420 nm (A420) was determined and the Miller Units of β-galactosidase activity were calculated as (A420)(1000)/(OD600)(t)(v) where t is the time in minutes and v is the volume of culture used in the assay in ml. The β-galactosidase activity of CB15 plac290 (empty vector control) was used as a blank sample reference.
c-di-GMP quantification.
Strains were grown to early-log growth phase (OD600 0.15–0.25) in the indicated media. 1 mL of culture was centrifuged for 5 min at 21,000 x g and the supernatant was removed by aspirating. Pellets were resuspended in 200 µL cold extraction buffer (1:1:1 mix of methanol, acetonitrile, and distilled H2O + 0.1 N formic acid) and incubated at −20 °C for 30 min. Samples were then centrifuged at 21,000 x g to pellet cell debris, and the supernatant was transferred to a fresh tube and stored at −80 °C until use. Experimental extraction solutions were desiccated overnight in a SpeedVac, re-solubilized in 100 µL of ultra-pure water, briefly vortexed, and centrifuged for 5 min at 21,000 x g to pellet insoluble debris. The clarified extract solutions were transferred to sample vials and analyzed by UPLC-MS/MS in negative ion-mode electrospray ionization with multiple-reaction monitoring using an Acquity Ultra Performance LC system (Waters Corp.) coupled with a Quattro Premier XE mass spectrometer (Waters Corp.) over an Acquity UPLC BEH C18 Column, 130Å, 1.7 µm, 2.1 mm X 50 mm. C-di-GMP was identified using precursor > product masses of 689.16 > 344.31 with a cone voltage of 50.0 V and collision energy of 34.0 V. Quantification of c-di-GMP in sample extracts was determined using a standard curve generated from chemically synthesized c-di-GMP (AXXORA). The standard curve solutions were prepared using 2-fold serial dilutions of c-di-GMP (1.25 µM – 19.0 nM) in ultra-pure water that were further diluted 1:10 into biological extracts (final c-di-GMP concentrations: 125 nM – 1.9 nM) from a low c-di-GMP strain of Caulobacter crescentus lacking several diguanylate cyclases (cdG0) described previously (Abel et al., 2013) which had been grown, harvested, extracted, dessicated and solubilized in ultra-pure water in tandem with the experimental samples described above. General UPLC buffer preparations, chromatographic gradients, and MS/MS parameters were performed using a previously published method (Severin & Waters, 2017). Intracellular concentrations of c-di-GMP were calculated using a previously published method (Massie et al., 2012) assuming C. crescentus average cellular dimensions of 6.46 × 10−16 l while the total number of cells present in each extraction were calculated by normalizing OD600 for each sample to the average CFUs found for WT cultures grown to OD600 0.2 (2 × 109 CFU/ml).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants R01GM102841 and R35GM122556 from the National Institutes of Health to YB, by National Science Foundation fellowship 1342962 to CE, by grant GM109259 from the National Institutes of Health to CW, and by National Institutes of Health Ruth L. Kirschstein National Research Service Award Predoctoral Fellowship F31 GM100770–01 to RM. We thank A Ducret for his help with the quantitative analysis of the holdfast delay experiments and critical reading of the manuscript, C Simmons for constructing YB4036 and YB4037, the Jenal and Thanbichler laboratories for providing strains and plasmids, and the members of the Brun laboratory for critical comments on the manuscript.
REFERENCES
- Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel Zur Wiesch P & Jenal U, (2013) Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the caulobacter cell cycle. PLoS Genet 9: e1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA & Jenal U, (2011) Regulatory Cohesion of Cell Cycle and Cell Differentiation through Interlinked Phosphorylation and Second Messenger Networks. Mol Cell 43: 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge P, Paul R, Goymer P, Rainey P & Jenal U, (2003) Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol Microbiol 47: 1695–1708. [DOI] [PubMed] [Google Scholar]
- Anderson PE & Gober JW, (2000) FlbT, the post‐transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol Microbiol 38: 41–52. [DOI] [PubMed] [Google Scholar]
- Belas R, (2014) Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22: 517–527. [DOI] [PubMed] [Google Scholar]
- Berne C, Ducret A, Hardy GG & Brun YV, (2015) Adhesins Involved in Attachment to Abiotic Surfaces by Gram-Negative Bacteria. Microbiol Spectr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne C, Ellison CK, Ducret A & Brun YV, (2018) Bacterial adhesion at the single-cell level. Nat Rev Microbiol. [DOI] [PubMed] [Google Scholar]
- Berne C, Kysela DT & Brun YV, (2010) A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm. Mol Microbiol 77: 815–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne C, Ma X, Licata NA, Neves BR, Setayeshgar S, Brun YV & Dragnea B, (2013) Physiochemical properties of Caulobacter crescentus holdfast: a localized bacterial adhesive. J Phys Chem B 117: 10492–10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair KM, Turner L, Winkelman JT, Berg HC & Kearns DB, (2008) A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320: 1636–1638. [DOI] [PubMed] [Google Scholar]
- Bodenmiller D, Toh E & Brun YV, (2004) Development of surface adhesion in Caulobacter crescentus. J Bacteriol 186: 1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Hardy GG, Trimble MJ & Brun YV, (2009) Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol 54: 1–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR & Lappin-Scott HM, (1995) Microbial biofilms. Ann Rev Microbiol 49: 711–745. [DOI] [PubMed] [Google Scholar]
- Donlan RM, (2002) Biofilms: microbial life on surfaces. Emerg Infect Dis 8: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, Quardokus EM & Brun YV, (2016) MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1: 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B & Jenal U, (2009) Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB & Brun YV, (2017) Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B, (1991) Genetics of Caulobacter crescentus. Methods Enzymol 204: 372–384. [DOI] [PubMed] [Google Scholar]
- Entcheva-Dimitrov P & Spormann AM, (2004) Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J Bacteriol 186: 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulds-Pain A, Birchall C, Aldridge C, Smith WD, Grimaldi G, Nakamura S, Miyata T, Gray J, Li G & Tang JX, (2011) Flagellin redundancy in Caulobacter crescentus and its implications for flagellar filament assembly. J Bacteriol 193: 2695–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH & Crosson S, (2014) A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet 10: e1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC & Wingender J, (2010) The biofilm matrix. Nat Rev Microbiol 8: 623–633. [DOI] [PubMed] [Google Scholar]
- Fong JN & Yildiz FH, (2015) Biofilm Matrix Proteins. Microbiol Spectr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritts RK, LaSarre B, Stoner AM, Posto AL & McKinlay JB, (2017) A Rhizobiales-Specific Unipolar Polysaccharide Adhesin Contributes to Rhodopseudomonas palustris Biofilm Formation across Diverse Photoheterotrophic Conditions. Appl Environ Microbiol 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Neubauer M, Yang A, Johnson N, Morse M, Li G & Tang JX, (2014) Altered motility of Caulobacter Crescentus in viscous and viscoelastic media. BMC microbiology 14: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goller CC & Romeo T, (2008) Environmental influences on biofilm development. Curr Top Microbiol Immunol 322: 37–66. [DOI] [PubMed] [Google Scholar]
- Hardy GG, Allen RC, Toh E, Long M, Brown PJ, Cole-Tobian JL & Brun YV, (2010) A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol 76: 409–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy GG, Toh E, Berne C & Brun YV, (2017) Mutations in sugar-nucleotide synthesis genes restore holdfast polysaccharide anchoring to Caulobacter crescentus holdfast anchor mutants. J Bacteriol: JB. 00597–00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Pérez M, Setayeshgar S, Hou Y, Temam R, Brun YV, Dragnea B & Berne C, (2018) Layered Structure and Complex Mechanochemistry Underlie Strength and Versatility in a Bacterial Adhesive. mBio 9: e02359–02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW & Harwood CS, (2008) Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69: 376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MD, Zucker LI, Brown PJ, Kysela DT, Brun YV & Jacobson SC, (2015) Timescales and Frequencies of Reversible and Irreversible Adhesion Events of Single Bacterial Cells. Anal Chem 87: 12032–12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry A, Briandet R, Aymerich S & Gohar M, (2010) Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 156: 1009–1018. [DOI] [PubMed] [Google Scholar]
- Hug I, Deshpande S, Sprecher KS, Pfohl T & Jenal U, (2017) Second messenger–mediated tactile response by a bacterial rotary motor. Science 358: 531–534. [DOI] [PubMed] [Google Scholar]
- Javens J, Wan Z, Hardy GG & Brun YV, (2013) Bypassing the need for subcellular localization of a polysaccharide export-anchor complex by overexpressing its protein subunits. Mol Microbiol 89: 350–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U & Malone J, (2006) Mechanisms of Cyclic-di-GMP Signaling in Bacteria. Annu Rev Genet 40: 385–407. [DOI] [PubMed] [Google Scholar]
- Johnson RC & Ely B, (1977) Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan E & Watnick P, (2009) Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73: 310–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA & Sternberg MJ, (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Kim TJ, Young BM & Young GM, (2008) Effect of flagellar mutations on Yersinia enterocolitica biofilm formation. Appl Environ Microbiol 74: 5466–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi A & Jenal U, (2006) Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol 188: 5315–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C & Brun YV, (2012) Surface contact stimulates the just‐in‐time deployment of bacterial adhesins. Mol Microbiol 83: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie JP, Reynolds EL, Koestler BJ, Cong J-P, Agostoni M & Waters CM, (2012) Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci USA 109: 12746–12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker RI & Smit J, (1988) Characterization of the Adhesive Holdfast of Marine and Freshwater Caulobacters. Appl Environ Microbiol 54: 2078–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt PM, Danhorn T & Fuqua C, (2007) Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J Bacteriol 189: 8005–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, NY. [Google Scholar]
- Muir RE & Gober JW, (2004) Regulation of FlbD activity by flagellum assembly is accomplished through direct interaction with the trans‐acting factor, FliX. Mol Microbiol 54: 715–730. [DOI] [PubMed] [Google Scholar]
- Ong CJ, Wong ML & Smit J, (1990) Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J Bacteriol 172: 1448–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter JS, (1964) Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev 28: 231–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A & Crosson S, (2007) A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci USA 104: 18241–18246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried JL & Collmer A, (1987) An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57: 239–246. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS & Eliceiri KW, (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin GB & Waters CM, (2017) Spectrophotometric and Mass Spectroscopic Methods for the Quantification and Kinetic Evaluation of In Vitro c-di-GMP Synthesis In: c-di-GMP Signaling. Springer, pp. 71–84. [DOI] [PubMed] [Google Scholar]
- Smith CS, Hinz A, Bodenmiller D, Larson DE & Brun YV, (2003) Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol 185: 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprecher KS, Hug I, Nesper J, Potthoff E, Mahi MA, Sangermani M, Kaever V, Schwede T, Vorholt J & Jenal U, (2017) Cohesive Properties of the Caulobacter crescentus Holdfast Adhesin Are Regulated by a Novel c-di-GMP Effector Protein. mBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Hsieh ML, Khataokar A, Neiditch MB & Waters CM, (2013) Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol 90: 1262–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todhanakasem T & Young GM, (2008) Loss of flagellum-based motility by Listeria monocytogenes results in formation of hyperbiofilms. J Bacteriol 190: 6030–6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh E, Kurtz HD Jr. & Brun YV, (2008) Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol 190: 7219–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolker-Nielsen T, (2015) Biofilm Development. Microbiol Spectr 3: MB-0001–2014. [DOI] [PubMed] [Google Scholar]
- Van Dellen KL, Houot L & Watnick PI, (2008) Genetic analysis of Vibrio cholerae monolayer formation reveals a key role for ΔΨ in the transition to permanent attachment. J Bacteriol 190: 8185–8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Brown PJ, Elliott EN & Brun YV, (2013) The adhesive and cohesive properties of a bacterial polysaccharide adhesin are modulated by a deacetylase. Mol Microbiol 88: 486–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J & Newton A, (1997) Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol Microbiol 24: 233–239. [DOI] [PubMed] [Google Scholar]
- Xu J, Kim J, Danhorn T, Merritt PM & Fuqua C, (2012) Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res Microbiol 163: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.