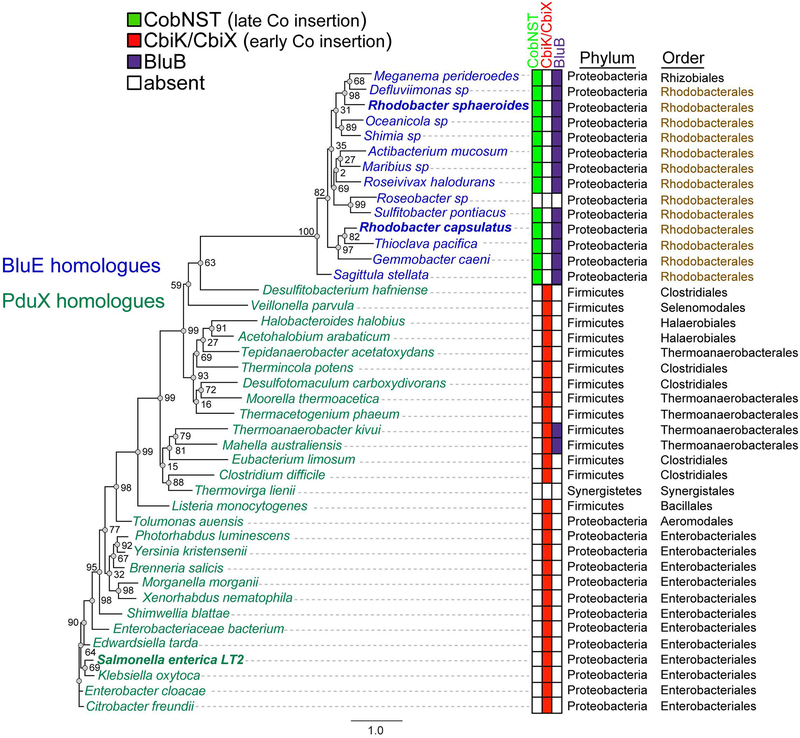
Figure 3. Phylogenetic analysis of the distribution of PduX and BluE proteins.
Maximum likelihood phylogenetic tree of homologous proteins based on the amino acid sequence of SePduX (dark green) and RsBluE or RcBluE (blue). Order Rhodobacterales is highlighted in brown. Color-coded table of the presence or absence of the cobaltochelatase CbiK or CbiX (red squares), which is indicative of the early-cobalt-insertion pathway, the presence of all three subunits of the cobaltochelatase CobNST (lime green squares) in organisms that use the late-cobalt-insertion pathway and BluB (purple squares) the O2-dependent DMB synthase, which is indicative of a physiological reliance on Cbas with DMB as the lower ligand base. Sequences were aligned using the MUSCLE (Edgar, 2004) plugin within Geneious R8.1.7 software (Biomatters Ltd.) with 100 iterations and default settings. Maximum likelihood phylogenic tree was generated with the online PhyML (Guindon et al., 2010) tool on the ATCG Montpellier Bioinformatics Platform available at http://www.atgc-montpellier.fr/phyml/, using Jones-Taylor-Thornton substitution model (Jones et al., 1992) with 500 bootstrap replicates. Bootstrap support for each node is shown as a percent value. The scale bar is provided as a reference for branch lengths. Gene locus tags are available in Table S1.