Fig. 2.
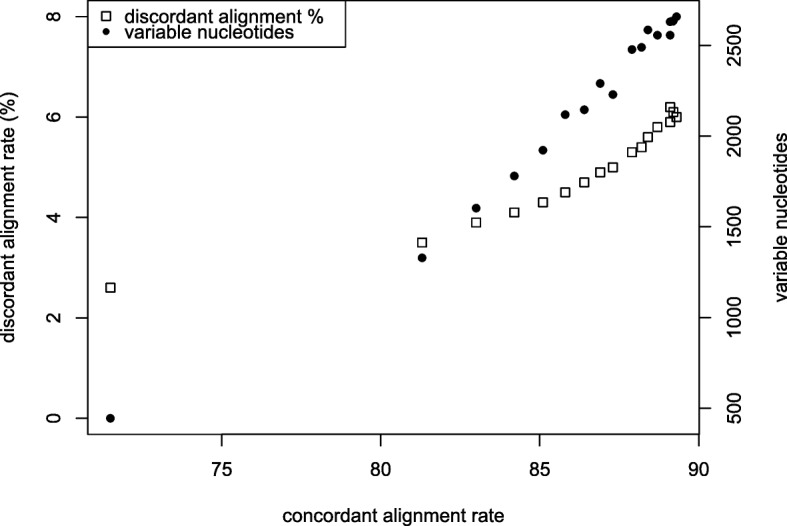
The effects of alignment parameters on alignment attributes. As the frequency of concordant read alignments increases (x-axis), the frequency of discordant paired end read alignments increases (left y axis) and the number of variable nucleotides at single genomic positions increases (right y-axis). Alignment criteria set nine, the criteria set used in this study, results in concordant frequency of 87.3%, discordant frequency of 5%, and 2229 variable nucleotides. The data is from sample 1 chromosome 1 alignments
