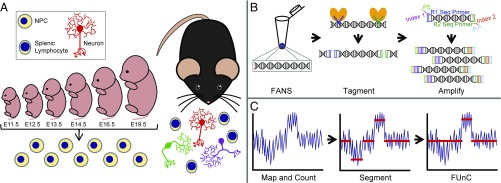
Fig. 1.
Overview of study design and methods. (A) An extensive range of murine tissue samples was collected for analysis, including NPCs at six embryonic ages, adult cortical neurons, and adult splenocytes. (B) The TbA method to amplify genomic DNA was performed on single nuclei isolated by FANS, which involves tagmentation—enzymatic DNA fragmentation via insertion of universal sequencing adapters—followed by PCR with unique sample indexes. (C) Bioinformatic processing of data begins with calculating sequencing depth in ∼0.1-Mb genomic regions, followed by CNV calling with the CBS algorithm, and finished with application of FUnC, which removes CNV calls that do not conform sufficiently to an integer copy number state.