Fig. 1.
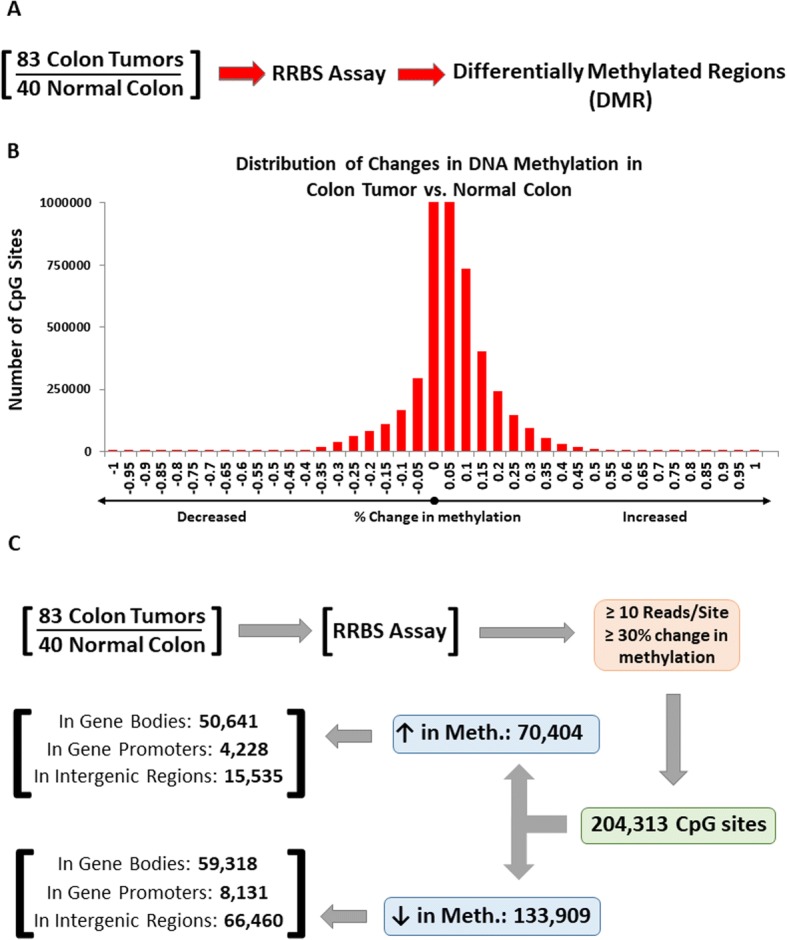
Genome-wide changes in DNA methylation in colon tumors. a Schematic of reduced representation bisulfite sequencing (RRBS) analysis workflow for 83 colon tumors vs. 40 normal colon samples. b Distribution of average change in DNA methylation in colon tumors vs. normal colon. c Distribution of differentially methylated CpG sites in colon tumors across the human genome. Using a stringent cutoff of > 30% change in DNA methylation, we identified over 204,000 CpG sites that are differentially methylated in colon tumors vs. normal colon. We further separated CpG sites that become hyper- vs. hypomethylated, and their genomic location (gene promoter, gene body, or intergenic region)
