Figure 6. Percentages of different subcellular structures associated with dig-pDNA.
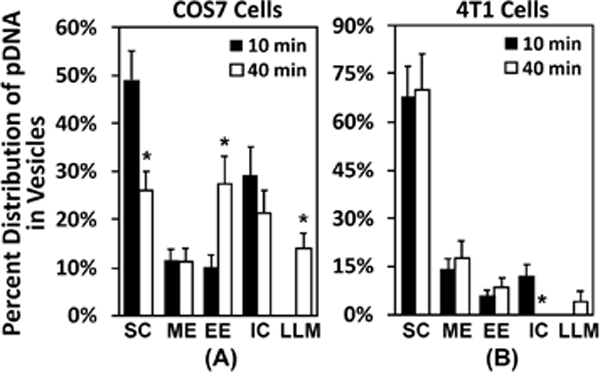
The raw data for this figure are the same as those for Fig. 5. Instead of reporting the total numbers, the figure shows the percentages of each subcellular structures that were associated with the EDS. The structures include surface complex (SC), macropinosome-like compartment (ME), early endosome-like compartment (EE), intracellular complex (IC), and late endosome-, lysosome-, or multivesicular body-like compartment (LLM). The details of the definition of each structure are provided in the results section. The data demonstrate that most pDNA molecules were located near the plasma membrane at 10 min. (A) In easy-to-transfect cells (COS7), a large fraction of pDNA was transferred to the inner vesicles at 40 min. (B) The transfer was minimal in hard-to-transfect cells (4T1). Bars and error bars represent the mean and the standard error of the mean, respectively. *p < 0.05, 10 min vs 40 min for each structure.
