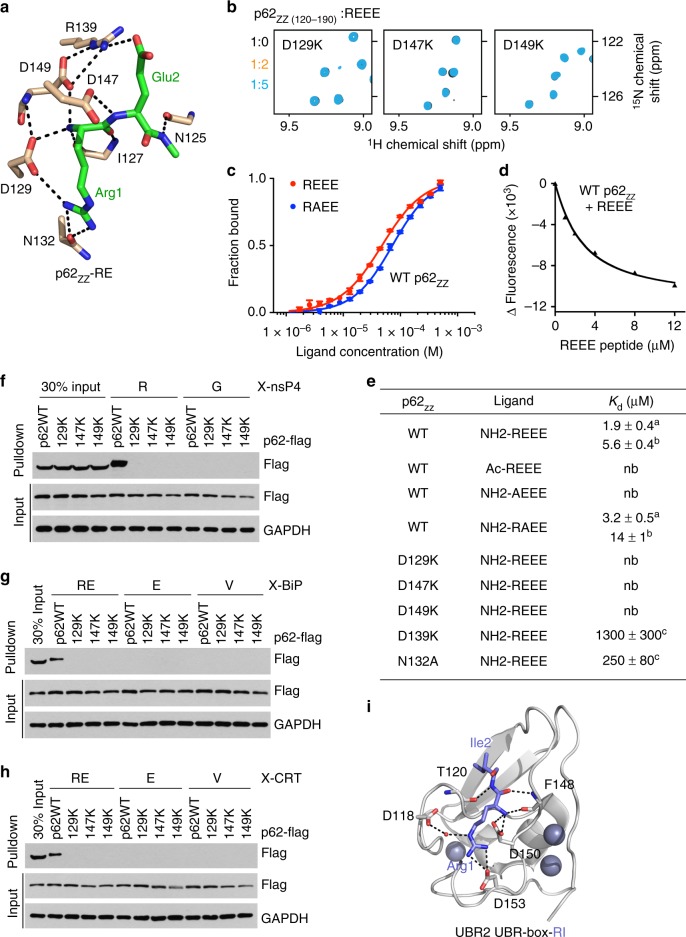
Fig. 2.
Molecular basis for the specific targeting of Nt-R by p62ZZ. a A zoom-in view of the Nt-R degron binding site. b Superimposed 1H,15N HSQC spectra of p62ZZ (120-190) collected upon titration with the REEE peptide. Spectra are color coded according to the protein:peptide molar ratio. c, d Representative binding curves used to determine the Kd values by MST (c) or tyrosine fluorescence (d). Error bar in c represents s.d. in triplicate measurements. e Binding affinities of p62ZZ for the indicated peptides measured by MST (a, using p62ZZ (115-190)) or tyrosine fluorescence (b, using p62ZZ (120-171)) or NMR (c, using p62ZZ (115-190)). Errors represent s.d. calculated from curve fitting (a, c) or duplicate midarguments (b). f–h Pulldown assays of wild-type or mutated p62-Flag expressed in HEK293 cells using X-nsP4 (f), X-BiP (g), and X-CRT (h) peptides. Uncropped blots are shown in Supplementary Figure 7. i A ribbon diagram of the structure of UBR-box of UBR2 (gray) in complex with Nt-R degron (blue, PDB 3NYN)