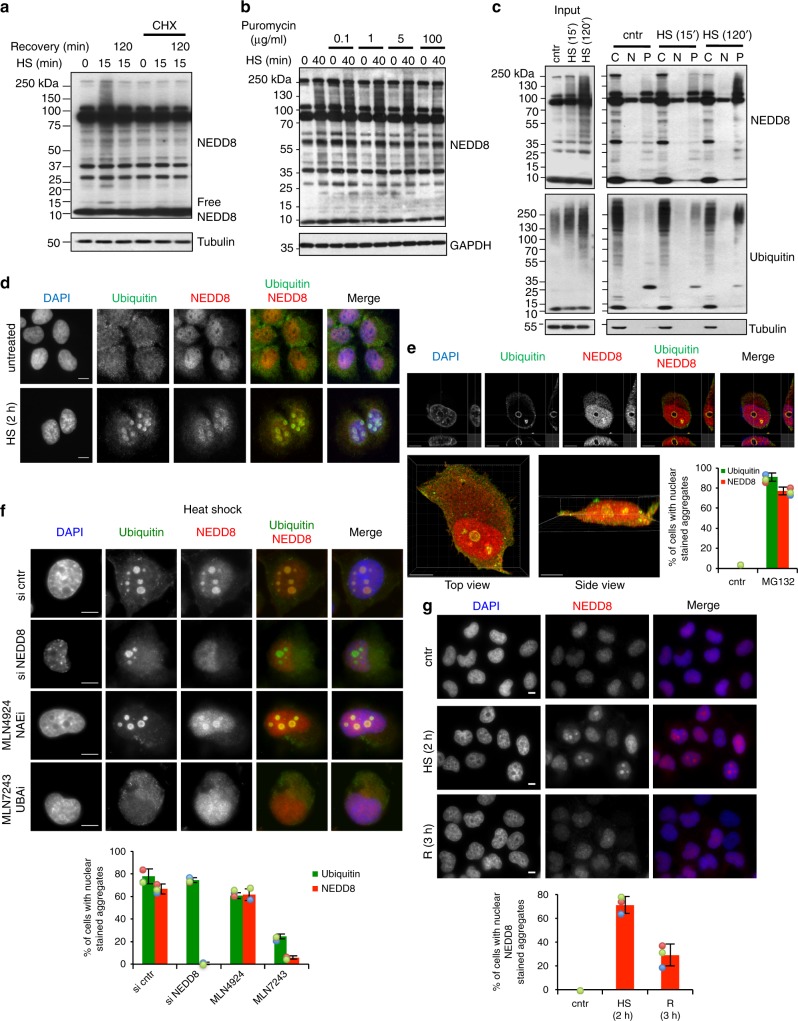
Fig. 1.
NEDD8 targets newly synthesised proteins and accumulates in nuclear aggregates upon proteotoxic stress. a U2OS cells were pretreated or not with Cycloheximide (CHX) at 100 µg/ml for 90 min, and then treated as indicated. After heat shock at 43 °C, cells were recovered at 37 °C for 2 h. b U2OS cells were pretreated or not with puromycin at the indicated concentrations for 30 min then heat-shocked. Cell extracts were analysed by western blotting. c U2OS cells were either untreated or heat-shocked at 43 °C for the indicated time. After HS, cells were subjected to subcellular fractionation as described in Methods (C, Cytoplasm; N, Nucleoplasm; P, Pellet) and fractions were analysed by western blotting. Tubulin was used as fractionation marker. d U2OS cells were either untreated or heat-shocked before staining for NEDD8 (red) and ubiquitin (green). Nuclei were stained with DAPI (blue). e Similar as in d, but cells were pretreated with MG132 (5 µM-overnight-ON). 3D rendering was performed as described in Methods. Top panel represents single plane images in different axis. Bottom images are the 3D reconstructions for the top and side view. Scale bar: 10 μm. Bottom right graph represents the % of cells with nuclear aggregates stained with NEDD8 or ubiquitin. Approximately 100 cells were counted per condition. f siRNA transfection in H1299 cells was performed as indicated for a total of 36 h. MLN4924 and MLN7243 were used at 500 nM for 15 and 5 h respectively before heat shock. Staining and quantitation of the experiment (bottom graph) were performed as above. g H1299 cells were heat-shocked as before and were allowed to recover at 37 °C as indicated. After fixation, cells were stained for NEDD8 (red) and DAPI (blue). Bottom graph represents quantitation of the experiment in (g), performed as above. For all bar graphs, values represent the average of three independent experiments ± standard deviation (SD). Scale bars, 10 µm