Abstract
Background
There is considerable discordance in the curve progression of adolescent idiopathic scoliosis (AIS) patients between monozygotic (MZ) twins, indicating that nongenetic factors must be involved in the curve progression of AIS patients. Epigenetic processes may constitute one of these factors and have not yet been investigated in relation to curve progression in AIS patients.
Methods
The genome and methylome of peripheral monocytes were compared between MZ twins discordant for curve progression. Sets of differentially methylated sites were validated using the MassARRAY platform of Sequenome on additional samples.
Results
In the discovery study, we found evidence suggesting a lack of differences at the genome sequence level and the presence of epigenetic differences related to the curve progression of AIS patients. The top 4 differentially methylated CpG sites associated with curve severity were tested, and only site cg01374129 (CpG site located at chr8:122583383, Hg19) was confirmed in two replication cohorts. The methylation levels of site cg01374129 were significantly lower in the progression group than in the nonprogression group. Cox regression analysis demonstrated that hypo-methylation of site cg01374129 was an independent prognostic factor for curve severity. Site cg01374129 methylation as a marker achieved a sensitivity of 76.4% and a specificity of 85.6% in differentiating between samples from patients with and without curve progression (AUC = 0.827; 95% CI: 0.780 to 0.876).
Conclusion
Increased curvature is associated with decreased methylation at site cg01374129. Our results indicate that methylation of site cg01374129 may therefore serve as a promising biomarker in differing between patients with and without curve progression.
Keywords: Adolescent idiopathic scoliosis, Progression, Whole genome sequencing, Methylation, Predictor
Abbreviations: AIS, adolescent idiopathic scoliosis; MZ, monozygotic; SNV, single-nucleotide variant; CNV, copy number variation; Indels, insertions and deletions; AUC, Area under the ROC curve; CI, confidence interval; ROC, receiver operating characteristic; HR, hazard ratio; GATK, Genome Analysis Toolkit
Research in context.
Evidence before this study
Adolescent idiopathic scoliosis (AIS) is believed to be a complex, multifactorial disease that results from the interaction of genetic and environmental factors. The etiopathogenesis of AIS curve progression still remains unknown. Considerable research has indicated that the methylation of specific genes may serve as a biomarker for disease diagnostic screening and prognostic prediction. Research on the etiopathogenesis of AIS curve progression from the standpoint of epigenetics has been scarcely reported. We mainly explored the value of DNA methylation in predicting curve progression in AIS patients.
Added value of this study
We performed the first analysis of DNA methylation differences for the etiology of AIS curve severity using a genome-wide approach and found evidence suggesting a lack of differences at the genome sequence level and the presence of epigenetic differences related to curve progression of AIS patients. Thus, we suggest that environmental factors, not genetic factors, may lead to the etiology of AIS curve progression by epigenetic changes.
Implications of all the available evidence
Epigenetic, but not genetic, factors play a substantial role in the progression of AIS. Hypo-methylation of site cg01374129 is an independent predictor of high-risk curve progression and is a promising marker for identifying patients with a high risk of progression. Future studies are needed to validate the capabilities of methylation in predicting curve progression.
Alt-text: Unlabelled Box
1. Background
Adolescent idiopathic scoliosis (AIS) affects approximately 1% to 3% of children aged 10 to 16 years [1]. However, compared to the overall prevalence of AIS, only up to 0.1% of this population requires surgery [2]. In most cases, once a curve has been noticed, the patient must be closely followed for the likelihood of progression. This seems reasonable for patients who are at greatest risk of curve progression. However, for those with a low risk of progression, this would increase unnecessary physician visits, cost and radiation exposure. Therefore, the ability to identify the patients who are at greatest risk of curve progression and to customize treatment for patients has become increasingly important [3].
AIS is believed to be a complex, multifactorial disease that results from the interaction of genetic and environmental factors [4]. Despite high concordance rates within pairs of monozygotic (MZ) twin diagnosed with AIS, there are often considerable curve severity differences within AIS-concordant MZ twins [[5], [6], [7]]. Additionally, bracing can decrease the progression of curves and alter the natural history of idiopathic scoliosis [8]. This evidence strongly implicates a role for nongenetic factors in curve severity. Therefore, during the initiation phase of AIS, genetics probably has a greater role than do environmental factors, whereas environmental factors probably make a greater contribution to curve progression than do genetics [9].
It is now widely accepted that epigenetic changes, which may reflect the interaction of genetic and environmental factors, are causal to many developmental disorders [10]. Of these epigenetic changes, DNA methylation has been studied intensively. Specific DNA methylation patterns ensure proper gene expression, thereby regulating various fundamental biological processes, such as cellular differentiation and development [11]. Considerable literature has indicated that the methylation of specific genes may serve as a biomarker for disease diagnostic screening and prognostic prediction [[12], [13], [14], [15], [16]]. Nevertheless, research on the etiopathogenesis of AIS curve progression from the standpoint of epigenetics has been scarcely reported [17].
In this research, we mainly focused on the value of DNA methylation in predicting curve progression in AIS patients. We first identified candidate differentially methylated loci by performing epigenomic analyses within MZ AIS twins discordant for curve severity. Then, we investigated the methylation status of these loci in sporadic AIS patients who progressed compared to those who did not. Lastly, our data demonstrated DNA methylation in blood cells was associated with significant prognostic value in the prediction of curve progression.
2. Materials and methods
2.1. Subjects and samples
Two female twin pairs diagnosed with AIS, discordant for curve progression, were recruited at the Department of Orthopedics, Shanghai Changzheng Hospital, Second Military Medical University.
A cohort of 92 subjects with a confirmed diagnosis of AIS was nested in two groups (progression group and nonprogression group) for DNA methylation studies (Cohort 1). The progression group included 50 patients who presented with a curve over 45° on the first clinic visit, and the nonprogression group included 42 patients who reached skeletal maturity without curve progression to over 30° on the first clinic visit. Skeletal maturity was defined as a documented Risser sign of 4 or 5.
Another prospective cohort of 276 unrelated subjects was retrospectively reviewed to determine the prognostic value of DNA methylation in curve progression (Cohort 2). The inclusion criteria were first presented, age 10 to 16 years, untreated, with a Risser sign of 2 or below, and a Cobb angle between 10 and 25°. These patients were regularly followed and observed or treated according to their skeletal maturity as well as the severity and progression rate of the curve. Patients who had a Cobb angle of 25° to 29° with a documented curve progression of at least 5° in two successive clinical follow-ups or who had a Cobb angle between 30° and 40° were recommended to wear a brace until skeletal maturity. Patients with a Cobb angle of over 40° were offered surgical correction.
The characteristics of participants in the studied cohorts are summarized in Table 1. This study was conducted in accordance with the tenets of the Declaration of Helsinki and was approved by the Institutional Review Boards of Shanghai Changzheng Hospital. Written informed consent was obtained from the patients or their guardians prior to the study.
Table 1.
Characteristics of participants in the studied cohorts.
| Characteristics |
Two discovery MZ pairs |
Replication cohort 1 |
Replication cohort 2 |
|||
|---|---|---|---|---|---|---|
| Progressiongroup(n = 2) | Non-progressiongroup(n = 2) | Progressiongroup (n = 50) | Non-progressiongroup (n = 42) | Progression group(n = 112) | Non-progressiongroup (n = 164) | |
| Age at first visit (years) | 14 | 14 | 13.7 ± 2.4 | 13.3 ± 1.9 | 14.1 ± 2.1 | 13.8 ± 2.7 |
| Female, n (%) | 2(100) | 2(100) | 37(74) | 30(71) | 85(75.9) | 126(76.8) |
| BMI (kg/m2) | 15.91 | 15.39 | 16.43 | 16.88 | 15.97 | 16.24 |
| Weight (kg) | 42.4 | 41.7 | 45.6 ± 7.2 | 44.9 ± 8.1 | 46.5 ± 6.6 | 45.8 ± 6.2 |
| Height (cm) | 163.2 | 164.6 | 163.4 ± 4.7 | 164.1 ± 5.5 | 165.4 ± 5.2 | 164.6 ± 5.1 |
MZ, monozygotic; BMI, body mass index.
Peripheral blood lymphocytes were collected for all of the recruited subjects. DNA samples were extracted from peripheral blood using a blood DNA extraction kit (QIAamp DNA Blood Mini Kit; Qiagen, Germany) according to the manufacturer's protocol. DNA samples were stored at −20 °C until use. DNA integrity was evaluated by 1% agarose gel electrophoresis.
2.2. Whole-genome sequencing
The sequencing was performed using the Applied BiosystemSOLiD4.0 (Sequencing by Oligonucleotide Ligation and Detection) system. The primary sequencing data were analyzed by using the standard SOLiD analysis workflow. The reads were aligned for single-nucleotide polymorphism (SNP) calling and subsequent analysis for prioritization of candidate genes. Detected sequence variants were removed if they were present in the dbSNP, HapMap, 1000 Genomes, ESP6500, ExAC, or in-house Chinese Exome Database (1500 Chinese Han individuals).
2.3. Whole-exome sequencing
Approximately 10 μg of DNA from one pair of monozygotic twins was sent to Genergy Biotech (Shanghai, China) for whole-exome sequencing (WES). A Nimblegen Exome V4 Kit (Roche) was used for exon capture. Exomes were then sequenced on an Illumina Hiseq3000 (Illumina) according to the manufacturer's instructions. UCSC Genome Browser build Hg19 was used as the reference sequence. Realignment and variant calling were performed as previously described. All SNVs and indels were subsequently filtered against data from the databases mentioned above. Deleterious SNVs were predicted by LRT, Polyphen-2, and MutationTaster programs.
2.4. Zygosity analysis
We evaluate relationship between individuals from two MZ pairs by estimating the probability of identity-by-descent (IBD). A full treatment of relatedness that allows for inbreeding requires a set of nine probabilities. Here, we assumed an absence of inbreeding, and we therefore needed to estimate only the three probabilities (Z2, Z1 or Z0) that corresponded to situations wherein two, one or zero pairs of the alleles were carried by two individuals at the same SNP and were IBD. These probabilities can be estimated quickly using an approximate moment method implemented in PLINK [18]. To further confirm that the twins were monozygotic, we used a short tandem repeat (STR) typing kit (AmpFLSTR™ Identifiler™ Plus PCR Amplification Kit; Applied Biosystems, Foster City), which amplifies 15 short tandem repeat loci and the amelogenin locus. DNA analysis was performed on an ABI 3730× DNA analyzer with GeneMapper5.0 software (Applied Biosystems, Foster City). Kinship analysis was not performed among patients from the two cohorts, as none of them reported a family history of scoliosis.
2.5. Illumina Infinium Human Methylation 850 analysis
The isolated DNA samples underwent bisulfite conversion using an EZ DNA Methylation-Gold™ kit (Zymo Research Corporation) and were then subjected to genome-wide DNA methylation profiling using the Illumina Infinium HumanMethylation850 BeadChip assay (Illumina, San Diego, CA), according to the manufacturer's protocol.
2.6. Quantitative DNA methylation
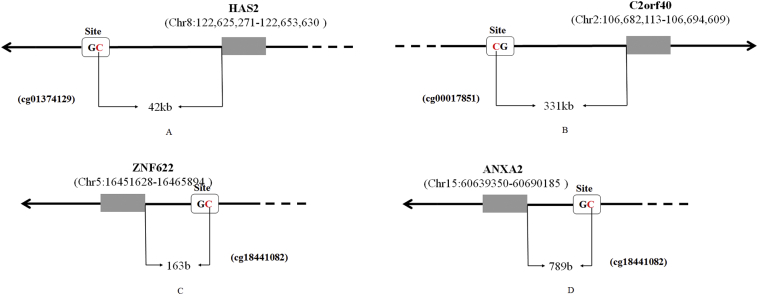
Quantitative methylation analysis of cg00017851 (CpG site located at chr2:106351195, Hg19), cg18441082 (CpG site located at chr5:16466057, Hg19), cg11421255 (CpG site located at chr15:60690974, Hg19), and cg01374129 (CpG site located at chr8:122583383, Hg19) (Fig. S1) was performed by the MassARRAY compact system (CapitalBio Beijing, China). DNA was extracted by a QIAamp DNA Mini Kit (QIAGEN). Then, the DNA sample was treated with sodium bisulfite (EZ DNA Methylation-Gold Kit, ZYMO) for conversion, and PCR was conducted for DNA amplification. The primer pairs are shown in Table S1. For each forward primer, an additional 10-mer tag was added for balance, and a T7 prompter was added for in vitro transcription. The PCR amplification that was conducted obeyed the following parameters: hot start at 94 °C for 10 min; followed by denaturing at 94 °C for 45 s, annealing at 62 °C for 48 s and decreasing by 0.5 °C per cycle, extension at 72 °C for 1 min for 10 cycles; then, denaturing at 94 °C for 45 s, annealing at 57 °C for 48 s, extension at 72 °C for 1 min for 35 cycles; and final incubation at 72 °C for 3 min. After amplification, unincorporated dNTPs were dephosphorylated (SAP treatment) by a MassCLEAVE Kit (SEQUENOM). The reaction mixture included 1.6 μl of premix and 4.0 μl PCR products. The reaction system was incubated at 37 °C for 20 min and then heat inactivated for 5 min at 85 °C. After SAP treatment, 2 μl product and 5 μl RNase system (MassCLEAVE Kit, SEQUENOM) were mixed for in vitro transcription, according to the manufacturer's instructions. After all the procedures described, we used a MassARRAY Nanodispenser RS1000 (SEQUENOM) to condition the samples and spot them on a 384-pad Spectro-Chip (SEQUENOM), followed by spectral acquisition on a MassARRAY Compact System (SEQUENOM). The SpectroCHIP was supported by matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF), and the resultant methylation calls were analyzed by EpiTYPER software v1.0 (SEQUENOM).
Fig. S1.
Schematic diagram shows the CpG position of cg01374129 (chr8:122583383, A), cg00017851 (chr2:106351195, B), cg18441082 (chr5:16466057, C), and cg11421255 (chr15:60690974, D) in the human genome (HG19).
2.7. Statistical analysis
Data were summarized using mean ± standard deviation and proportions for continuous and categorical variables. The Mann-Whitney U test was applied to test for differences in DNA methylation levels between the two groups.
Progression-free survival was defined as the time between the first visit and the last visit before treatment or the date of last clinical visit and was used as the primary endpoint for outcome analysis. The Kaplan-Meier analysis and Cox proportional-hazards models were used to estimate the survival distributions. Univariate and multivariate Cox regression analyses were performed to identify the prognostic values of curve magnitude at presentation, curve type (single thoracic, thoracolumbar, single lumbar, double thoracic, and double lumbar), sex, age at presentation, Risser sign, menarche status (only for females), body mass index, and DNA methylation status. The area under the receiver operating characteristic (ROC) curve (AUC) was calculated, and 95% confidence intervals (CIs) were reported. All analyses were two sided, and P-values of <0.05 were regarded as significant.
Statistical calculations were performed using the statistical package SPSS version 17.0 for Windows (IBM Corp., Armonk, NY, USA) and GraphPad Prism version 6.00 for Windows (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. Identification and recruitment of MZ AIS twins
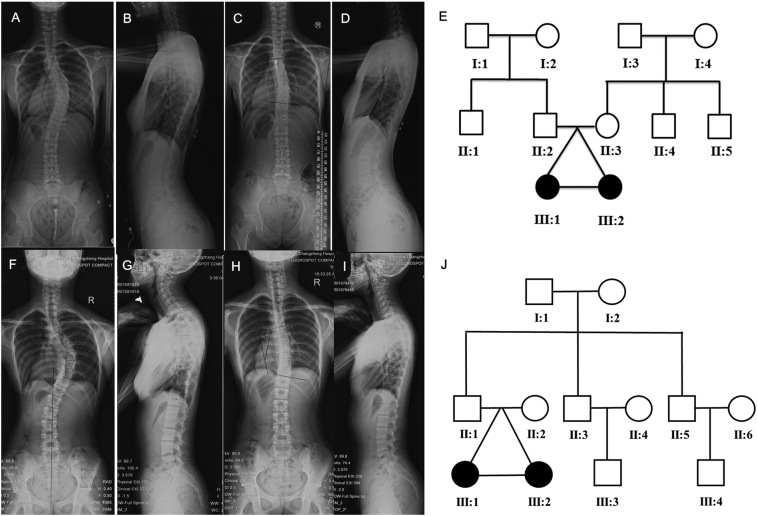
Two pairs of monozygotic twins (Pair 1 and Pair 2) from the Chinese Han population were recruited in our study (Fig. 1, Fig. 2). We assigned Pair 1A and Pair 2A as the older sisters from each MZ pair and Pair 1B and Pair 2B as the younger ones (Fig. 2). All four patients were diagnosed with AIS through history and physical examinations, including a complete neurological examination and standing full-spine posterior-anterior/lateral radiographs. AIS is defined as a lateral curvature of the spine >10° with vertebral rotation and no congenital deformity. Both Pair 1A and Pair 2A had a main thoracic curve with Cobb angle over 45° and had under gone surgery at our institution. Pair 1B and Pair 2B had much smaller curves of approximate 20° and had close clinical follow-ups. Their curves were <30°, even after skeletal maturity (Fig. 2). Both MZ pairs had identical demographic characteristics and highly similar environments.
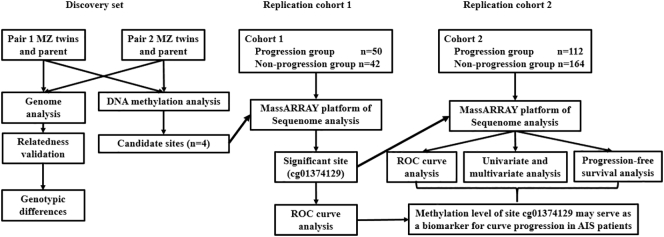
Fig. 1.
Overview of the study design.
Fig. 2.
Two pairs of monozygotic twins in the discovery study. (A and B) Posteroanterior and lateral views of individual Pair 1A show a main thoracic curve of 47° and thoracic kyphosis of 13°. (C and D) Posteroanterior and lateral views of individual Pair 1B show a main thoracic curve of 22° and thoracic kyphosis of 27°. (E) Pedigree and segregation pattern in the Pair 1 family. (F and G) Posteroanterior and lateral views of individual Pair 2A show a main thoracic curve of 50° and thoracic kyphosis of 10°. (H and I) Posteroanterior and lateral views of individual Pair 2B shows a main thoracic curve of 21° and thoracic kyphosis of 28°. (J) Pedigree and segregation pattern in the Pair 2 family. Solid symbols indicated affected individuals. Squares signify males and circles, females.
To confirm that both pairs were monozygotic, we estimated probabilities of IBD and analyzed short tandem repeat (STR) loci. The results showed that Pair 1A/1B and Pair 2A/2B had the same alleles, strongly suggesting that they were both monozygotic twins (Fig. S2 for IBD estimates and Table S2 for STR).
Fig. S2.
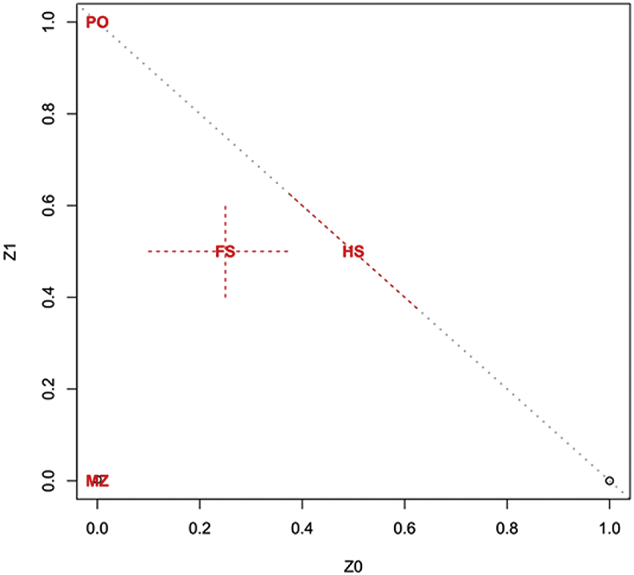
Relationship confirmation from IBD estimates for twins in the discovery study. Estimates of the IBD coefficients, Z0 and Z1, are used to infer relatedness. Each point is for a pair of samples and the diagonal line is Z0 + Z1 = 1. Twin pairs are expected to occur at MZ area at the lower left corner of the diagonal (Z0 = Z1 = 0). Unrelated pairs are expected to occur at the lower right end of the diagonal.
3.2. Genetic variants
For Pair 1, two genomes of blood cell-derived DNA were sequenced, resulting in 39.35× and 39.06× average coverage (Table S3). The discordant SNVs and Indels from GATK (Genome Analysis Toolkit) resulted in 6317 variants. After excluding nonexonic and synonymous variants, 637 SNVs, but no indels, were obtained from the whole-genome sequencing data. We then chose variants predicted to be potentially deleterious and disease-related by SIFT and by PolyPhen2 in our dataset. No discordant SNVs or indels were detected. Previous genome-wide association studies (GWAS) have identified many scoliosis-causing SNPs in noncoding genomic regions. We then further analyzed discordant variants in intronic and intergenic regions (Table S4). None were found to be in the list, which included previously identified genes associated with susceptibility and progression of AIS (Previous identified genetic markers shown in Table S5). From Pair 2, blood samples were exome sequenced. The exome sequencing dataset had average depths of 118.69 and 105.66 for Pair 2A and Pair 2B, respectively. Exome sequencing of both twins resulted in a total of 186,257 variants prior to the discordant analysis. After applying the stringent filtering criteria, no putative discordant SNVs or indels were detected. Another way that genomes can differ is through copy number variants (CNVs). Analysis of sequencing results did not suggest any presence of bona fide CNVs between the twins.
3.3. Genome-wide patterns of DNA methylation in MZ twins
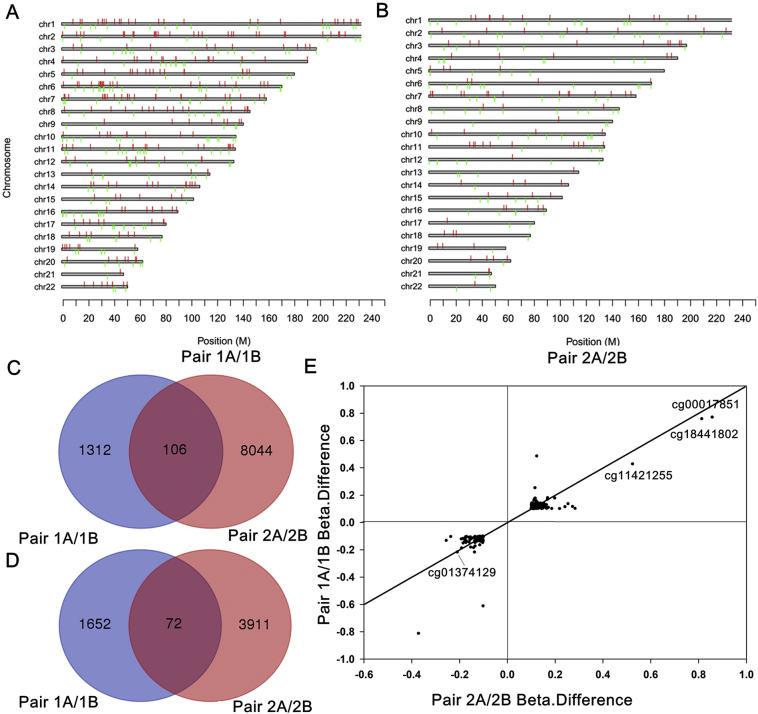
We performed a genome-wide DNA methylation study in blood samples from both MZ pairs using Illumina Infinium MethylationEPIC (850 K) arrays. Applying the thresholds of Δβ > |0.10| and P value <.05, we found 178 CpG sites to be differentially methylated when comparing twins with large curves with twins with small curves (Fig. 3A-D). The top 10 differentially methylated CpG sites associated with curve progression identified in monozygotic twins were presented in Table 2. We chose the top four differentially methylated sites with similar Δβ in both pairs for further analysis (Fig. 3E, Fig. S1). Among them, cg01374129 was negatively associated with curve progression and the other 3 (cg00017851, cg18441082, and cg11421255) were positively associated with curve progression.
Fig. 3.
Genome-wide methylation analyses in the discovery study. (A-B) The distribution of intra-pair differentiated methylation sites in various chromosomes. (C) Venn diagram showing the overlaps (middle) of intra-pair hypermethylated CpG sites. (D) Venn diagram showing the overlaps of intra-pair hypo-methylated CpG sites. (E) Plotting of the difference of beta values of the methylation sites in Pair 1A versus 1B (Pair 1A/1B) against that in Pair 2A versus 2B (Pair 2A/2B). Pearson correlation, r = 0.91 (P < .0001). Each symbol represents a methylation site. Pair 1A and 2A represent the individuals with large curves, and Pair 1B and 2B represent the individuals with small curves.
Table 2.
Top 10 differentially methylated CpG sites associated with curve progression identified in monozygotic twins.
| CpG | Position | P-value | Betadifference | Gene | Distance |
|---|---|---|---|---|---|
| cg00017851 | Chr2:106351195 | 0.002 | 0.813 | – | – |
| cg18441082 | Chr5:16466057 | 0.001 | 0.784 | ZNF622 | TSS200 |
| cg11421255 | Chr12:60690974 | 0.010 | 0.478 | ANXA2 | TSS1500 |
| cg10417124 | Chr2:102744380 | 0.044 | 0.196 | IL1R1 | 5’UTR |
| cg03671360 | Chr3:180297596 | 0.005 | 0.170 | – | – |
| cg11884832 | Chr6:167631788 | 0.009 | −0.142 | – | – |
| cg21983484 | ChrX:49687062 | 0.042 | −0.160 | CLCN5 | TSS200 |
| cg23146833 | Chr3:171175866 | 0.001 | −0.182 | TNIK | Body |
| cg24848599 | Chr1:228604800 | 0.000 | −0.185 | TRIM17 | TSS1500 |
| cg01374129 | Chr8:122583383 | 0.003 | −0.214 | – | – |
TSS200, 0-200 bp upstream of transcriptional start site; TSS1500, 201-1500 bp upstream of transcriptional start site.
3.4. Validation of differentially methylated sites using the MassARRAY platform of sequenome
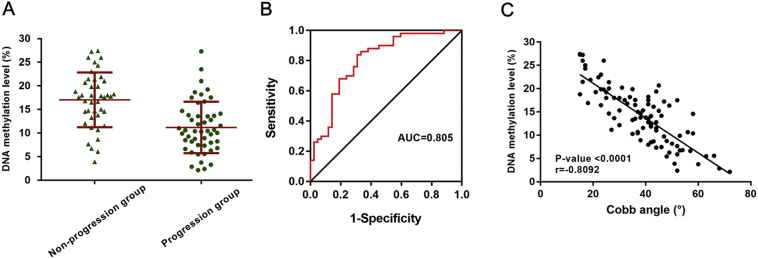
In validation Cohort 1, we measured the top four differentially methylated sites, including cg01374129, cg00017851, cg18441082 and cg11421255 (Fig. S1). The methylation levels of cg01374129 in progression AIS patients' blood samples were markedly lower than those in nonprogression AIS patients' blood samples (P < .0001, Fig. 4A). The ROC curve analysis for progression AIS, compared to nonprogression AIS, revealed that the methylation levels of cg01374129 had a high AUC value of 0.805 (95% CI 0.714–0.896) (Fig. 4B). The sensitivity and specificity for progression AIS of cg01374129 methylation level at a cut-off level of 14.6% were 69% and 84%, respectively. More interesting, linear correlation showed that methylation level of the site cg01374129 was negatively correlated with curve magnitude (n = 92, r = −0.8092, P < .0001) (Fig. 4C).
Fig. 4.
Replication study in Cohort 1. (A) The methylation levels of site cg01374129 were significantly lower in the progression group (n = 50) than in the nonprogression group (n = 42). (B) Receiver operating characteristic (ROC) curves for methylation levels of site cg01374129 in Cohort 1. (C) Correlation of methylation status and Cobb angle of patients in Cohort 1. Dot plot showing correlation between the methylation biomarkers (site cg01374129) and Cobb angle. The r-value represents the square of Pearson correlation coefficient.
3.5. Analysis of the association between cg01374129 methylation and clinical characteristics of AIS
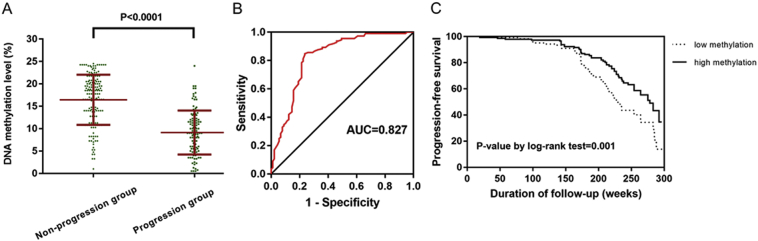
The predictive value of cg01374129 methylation levels was also validated in another cohort (Cohort 2) consisting of 276 patients. For these patients, the outcome variable was the time from presentation to the development of a curvature of at least 30° before any treatment [19,20]. A curvature of at least 30° was used to define curve progression because curves beyond 30° continue to progress even after skeletal maturity [21], but those <30° at skeletal maturity are much less likely to progress [[22], [23], [24]]. The methylation level of site cg01374129 was significantly lower in the progression group (curvature of at least 30°) than in the nonprogression group (<30° at skeletal maturity) (Fig. 5A). The ROC curve analysis showed that the AUC value was 0.827 (95% CI: 0.780 to 0.876), with a sensitivity of 76.4% and a specificity of 85.6% at a cut-off level of 15.1% (Fig. 5B).
Fig. 5.
Replication study in Cohort 2. (A) The methylation level of site cg01374129 was significantly lower in the progression group (n = 112) than in the nonprogression group (n = 164). (B) ROC curves for methylation levels of site cg01374129 in Cohort 2. (C) Kaplan-Meier curves of progression-free survival according to methylation levels of site cg01374129.
Data on progression-free survival were available for all 276 patients. Median follow-up was 168 weeks (range 15–298 weeks). Univariate survival analysis revealed that curve magnitude at presentation (P = .001, HR = 1.101, 95% CI = 1.042–1.163) and methylation status of site cg01374129 (P < .001, HR = 0.782, 95% CI = 0.731–0.837) were of prognostic significance (Table 3). On multivariate analysis, only the methylation level of site cg01374129 proved to be an independent prognostic factor associated with curve progression (high versus low, P < .001, HR = 0.782, 95% CI = 0.731–0.837). Moreover, Kaplan-Meier analysis revealed that progression-free survival was significantly impacted by methylation status of site cg01374129 (P = .001) (Fig. 5C).
Table 3.
Results from univariate analyses (Cox proportional hazard models).
| Variables | Unadjusted HR (95% CI) | P-value |
|---|---|---|
| Sex (female vs. male) | 1.237(0.526–2.908) | 0.625 |
| Curve magnitude (°) | 1.101(1.042–1.163) | 0.001 |
| Menarche status (post vs. pre) | 0.672(0.412–1.097) | 0.112 |
| Risser sign (≤2 vs. >2) | 1.052(0.636–1.739) | 0.844 |
| Location of the curve | ||
| Single thoracic | Reference | |
| Thoracolumbar | 1.250(0.555–2.815) | 0.590 |
| Single lumbar | 1.149(0.499–2.644) | 0.744 |
| Double thoracic | 1.937(0.882–4.258) | 0.100 |
| Double lumbar | 1.125(0.511–2.479) | 0.770 |
| BMI (≤24 vs. >24) | 0.953(0.583–1.557) | 0.847 |
| Methylation for cg01374129 (high vs. low) | 0.782(0.731–0.837) | <0.001 |
BMI, body mass index; HR, hazard ratio; CI, confidence interval.
4. Discussion
Several clinical parameters, including age, sex, curve magnitude, skeletal maturity, and pubertal status at presentation, have been conventionally used for predicting AIS curve progression, and researchers have developed formulae to calculate the risk of progression in individual patients based on these parameters [20,25]. However, the results are not very reliable [26]. Additionally, several SNPs were reported to be associated with curve progression, but the results had poor repeatability. Recently, Ogura et al. conducted the first GWAS for AIS progression and identified a functional variant, rs35333564 in MIR4300HG, that was related to curve progression in silico and in vitro [27]. Their results provided convincing evidence that this genetic factor is related to AIS curve progression and helped to improve the prediction method. In the present research, sequencing of the genome of the twins revealed no difference in the reported genetic marker, which led us to explore other possible mechanisms causing the discordant curve severity. In recent years, epigenetic alterations, such as aberrant DNA methylation patterns, have been extensively studied across multiple human diseases, with AIS being no exception [28].
To our knowledge, this research constitutes the first analysis of DNA methylation differences for the etiology of AIS curve severity using a genome-wide approach. It documents the presence of epigenetic differences (differential methylation) in MZ twin pairs discordant for curve severity, as well as between sporadic individuals with large curves and those with small curves. Thus, we suggest that environmental factors, not genetic factors, may lead to the etiology of AIS curve progression by epigenetic changes.
Our study has several important findings. First, we provide evidence suggesting a lack of genetic differences and the presence of epigenetic differences in MZ pairs who were discordant for curve magnitude. The use of disease-discordant MZ twins represents a powerful strategy in epigenetic epidemiology because identical twins theoretically have identical genome sequences, and recent studies have uncovered considerable epigenetic variations between MZ twins [[29], [30], [31], [32]]. In the discovery study, we performed next generation sequencing in two pairs of MZ twins. A rigorous discordant screening revealed no SNVs, Indels or CNVs potentially causing the twins' discordance. These results are in line with recent genetic research demonstrating that genetic differences between MZ twins, even those with discordant phenotypes, are very rare [29,33]. Therefore, we performed genome-wide DNA methylation analyses and found considerable intra-pair variation in methylation. The following replication studies confirmed a specific association between AIS curve progression and methylation status of site cg01374129 in blood cell DNA. Second, although MZ twins are matched for population cohort effects and exposure to many shared environmental factors, the observed DNA methylation differences highlight the role of nonshared environmental and stochastic factors in AIS curve progression. These findings are consistent with mounting data suggesting that environmentally mediated effects on the epigenome may be relatively common and important for disease [32,34]. Third, we demonstrate the significant correlation between DNA methylation and AIS curve magnitude across our sample cohort and highlight the importance of using epigenetic biomarkers as a predictor for curve severity. Site cg01374129 is located near hyaluronan synthase 2 (HAS2), which encodes putative hyaluronan synthases (HAS). Hyaluronan (hyaluronic acid, HA) is an important structural element distributed widely throughout all tissues of the skeletal system, and its synthesis occurs at the plasma membrane of cells via membrane-bound HAS [35,36]. Roughley analyzed the spine development in mice deficient in HAS2 gene expression and found that HA production by HAS2 was essential for normal vertebral and intervertebral disc development. Additionally, the absence of HAS2 impaired the organization of both soft and hard tissue elements within the spine [37]. It has previously been reported that AIS patients exhibit altered vertebral growth and wedging of the intervertebral disc [38,39]. Several researchers have suggested that abnormal spinal bone tissue is a key determinant of curve progression [[40], [41], [42]]. In the current study, hypo-methylation was found to be significantly associated with larger curves. We speculate that hypo-methylation of site cg01374129 might deregulate the expression of HAS2, impairing normal spine development and causing scoliosis progression.
This study has several strengths. First, our unique sample consisted of MZ twin pairs discordant for curve severity, as well as age-matched sporadic patients. Second, to our knowledge, this study is the first using a genome-wide approach to uncover differentially methylated loci that are associated with AIS curve progression. Third, although it was a small discovery cohort, we have excluded the possible influence of inherently infrequent genomic events.
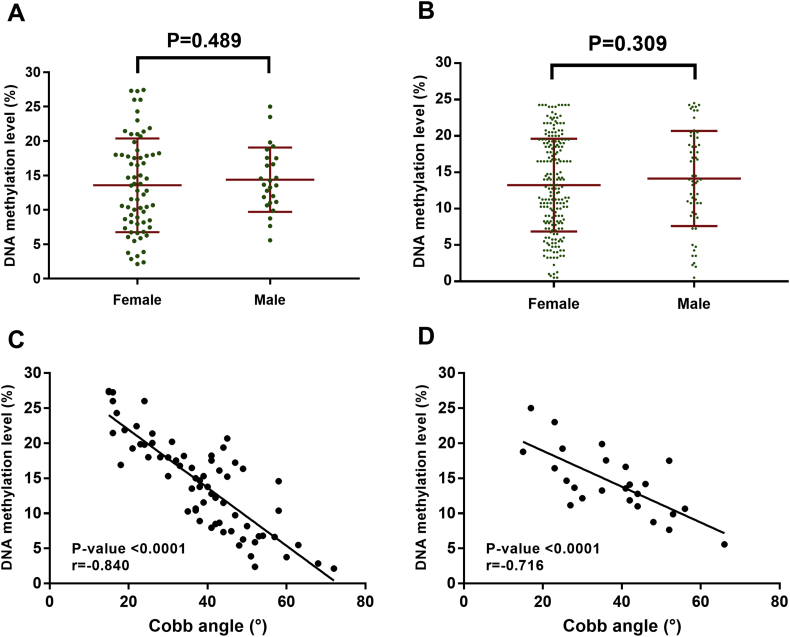
This study also has a number of limitations that should be considered when interpreting the results. First, the sample size for the discovery cohort is small, in part because MZ twins discordant for AIS curve magnitude are rare. Although our study revealed genomic similarity and epigenetic differences between the twins, other effects could also contribute to phenotype diversity. In addition, only 92 and 276 patients were recruited in the following replication study. Therefore, replication studies with larger samples are needed. Second, in the current study, the methylation status of site cg01374129 achieved a sensitivity of approximately 70%, with 85% specificity in differentiating between blood cells DNA from patients with curve progression and patients without curve progression. The addition of other prognostic tests, such as ScoliScore (Transgenomic), could probably increase the test sensitivity [3]. Third, methylation is a dynamic process influenced by environmental factors. Bracing has been proved to stop the progression of scoliosis and alter the natural history of scoliosis [8]. For patients in Cohort 2 initiating brace treatment due to a curve progressed over 30°, we did not compare the methylation status before and after brace-wearing. This issue should be investigated in future prospective studies in order to explain the mechanism of bracing at the molecular level. Forth, somatic mosaicism may play a role in the disease development, but in this research, we used only blood cell DNA for sequencing. In the future, multiple tissues, including skin fibroblasts and intervertebral disc tissue, can be used for the genetic investigations of mosaicism. Fifth, some authors have reported that the curvature of a female is more likely to progress than that of a male. We did not find this correlation in the present research. The results showed that methylation status of site cg01374129 was slightly lower in females than in males, but the difference was not statistically significant in both cohorts (Fig. S3 A and B). Linear correlation showed that methylation level of the site cg01374129 was negatively correlated with curve magnitude in both females (n = 67, r = −0.840, P < .0001) and males (n = 25, r = −0.716, P < .0001) in Cohort 1 (Fig. S3 C and D). These results collectively suggested that methylation status played a similar role in curve progression for females and males. Additionally, univariate and multivariate Cox regression analyses revealed that sex had no relationship with curve progression (Table 1 and Table 3). This finding was in consistent with those of Lonstein et al. [43] and Lara et al. [44] but should be interpreted with caution. It is possible that sex is not a risk factor of progression for Chinese females as it is in other populations. Therefore, the role of sex in curve progression should be further investigated in larger cohort enrolling patients with different racial background in a prospective design.
Fig. S3.
Methylation status of site cg01374129 stratified by sex in Cohort 1 (A) and Cohort 2 (B). Correlation of methylation status and Cobb angle in females (C) and males (D) in Cohort 1. Dot plot showing correlation between the methylation biomarkers (site cg01374129) and Cobb angle. The r-value represents the square of Pearson correlation coefficient.
In conclusion, this is the first analysis of DNA methylation differences for the etiology of AIS curve severity using a genome-wide approach. In this study, we found that the methylation status of site cg01374129, which is located near HAS2, has a significant negative correlation with AIS curve severity. In addition, our results indicate that hypo-methylation of site cg01374129 is an independent predictor of high-risk curve progression. Furthermore, we highlight the use of this promising marker in identifying patients with a high risk of progression. Future studies are needed to validate the capabilities of methylation in predicting curve progression.
The following are the supplementary data related to this article.
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
Ethics approval and consent to participate
The ethical committee and institutional review board of Shanghai Changzheng Hospital approved this study.
Funding
This research was supported by grants from Translational Medicine Centre of Second Military Medical University for Precision Medicine (2017JZ25) and the National Natural Science Foundation of China (81772305) to XZ.
Authors' contributions
YM, TL and SL made substantial contributions to the conception and coordination of the study and to acquisition and analysis of data. RG, SL and TL performed clinical work and data analysis. HJ and WS performed sample collection and the experiments. XZ and FY made substantial contributions to the study design, data interpretation, and study supervision. All authors critically revised the manuscript and gave final approval of the version to be submitted.
Acknowledgments
Acknowledgements
None.
Competing interests
None declared.
Contributor Information
Fu Yang, Email: yangfusq1997@smmu.edu.cn.
Xuhui Zhou, Email: zhouxuhui@smmu.edu.cn.
References
- 1.Weinstein S.L., Dolan L.A., Cheng J.C., Danielsson A., Morcuende J.A. Adolescent idiopathic scoliosis. Lancet. 2008;371(9623):1527–1537. doi: 10.1016/S0140-6736(08)60658-3. [DOI] [PubMed] [Google Scholar]
- 2.Lonstein J.E. Scoliosis: surgical versus nonsurgical treatment. Clin. Orthop. Relat. Res. 2006;443:248–259. doi: 10.1097/01.blo.0000198725.54891.73. [DOI] [PubMed] [Google Scholar]
- 3.Roye B.D., Wright M.L., Matsumoto H., Yorgova P., McCalla D., Hyman J.E. An Independent Evaluation of the Validity of a DNA-Based Prognostic Test for Adolescent Idiopathic Scoliosis. J. Bone Joint Surg. Am. 2015;97(24):1994–1998. doi: 10.2106/JBJS.O.00217. [DOI] [PubMed] [Google Scholar]
- 4.Miller N.H., Mims B., Child A., Milewicz D.M., Sponseller P., Blanton S.H. Genetic analysis of structural elastic fiber and collagen genes in familial adolescent idiopathic scoliosis. J. Orthop. Res. 1996;14(6):994–999. doi: 10.1002/jor.1100140621. [DOI] [PubMed] [Google Scholar]
- 5.Kesling KL, Reinker KA. Scoliosis in twins. A meta-analysis of the literature and report of six cases. Spine (Phila Pa 1976). 1997;22(17):2009–14; discussion 15. [DOI] [PubMed]
- 6.Hermus J.P., van Rhijn L.W., van Ooij A. Non-genetic expression of adolescent idiopathic scoliosis: a case report and review of the literature. Eur. Spine J. 2007;16(Suppl. 3):338–341. doi: 10.1007/s00586-007-0335-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simony A., Carreon L.Y., Hjmark K., Kyvik K.O., Andersen M.O. Concordance Rates of Adolescent Idiopathic Scoliosis in a Danish Twin Population. Spine (Phila Pa 1976) 2016;41(19):1503–1507. doi: 10.1097/BRS.0000000000001681. [DOI] [PubMed] [Google Scholar]
- 8.Weinstein S.L., Dolan L.A., Wright J.G., Dobbs M.B. Effects of bracing in adolescents with idiopathic scoliosis. N. Engl. J. Med. 2013;369(16):1512–1521. doi: 10.1056/NEJMoa1307337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng J.C., Castelein R.M., Chu W.C., Danielsson A.J., Dobbs M.B., Grivas T.B. Adolescent idiopathic scoliosis. Nat Rev Dis Primers. 2015;1 doi: 10.1038/nrdp.2015.30. [DOI] [PubMed] [Google Scholar]
- 10.Jones P.A., Laird P.W. Cancer epigenetics comes of age. Nat. Genet. 1999;21(2):163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 11.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 12.Su H.Y., Lai H.C., Lin Y.W., Chou Y.C., Liu C.Y., Yu M.H. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int. J. Cancer. 2009;124(2):387–393. doi: 10.1002/ijc.23957. [DOI] [PubMed] [Google Scholar]
- 13.Lind G.E., Guriby M., Ahlquist T., Hussain I., Jeanmougin M., Soreide K. Prognostic relevance of an epigenetic biomarker panel in sentinel lymph nodes from colon cancer patients. Clin. Epigenetics. 2017;9:97. doi: 10.1186/s13148-017-0397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meder B., Haas J., Sedaghat-Hamedani F., Kayvanpour E., Frese K., Lai A. Epigenome-Wide Association Study Identifies Cardiac Gene Patterning and A Novel Class of Biomarkers for Heart Failure. Circulation. 2017;136(16):1528–1544. doi: 10.1161/CIRCULATIONAHA.117.027355. [DOI] [PubMed] [Google Scholar]
- 15.D'Aquila P., Montesanto A., Mandala M., Garasto S., Mari V., Corsonello A. Methylation of the ribosomal RNA gene promoter is associated with aging and age-related decline. Aging Cell. 2017;16(5):966–975. doi: 10.1111/acel.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross-Innes C.S., Chettouh H., Achilleos A., Galeano-Dalmau N., Debiram-Beecham I., MacRae S. Risk stratification of Barrett's oesophagus using a non-endoscopic sampling method coupled with a biomarker panel: a cohort study. Lancet Gastroenterol Hepatol. 2017;2(1):23–31. doi: 10.1016/S2468-1253(16)30118-2. [DOI] [PubMed] [Google Scholar]
- 17.Burwell R.G., Dangerfield P.H., Moulton A., Grivas T.B. Adolescent idiopathic scoliosis (AIS), environment, exposome and epigenetics: a molecular perspective of postnatal normal spinal growth and the etiopathogenesis of AIS with consideration of a network approach and possible implications for medical therapy. Scoliosis. 2011;6(1):26. doi: 10.1186/1748-7161-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan K.J., Moe M.M., Vaithinathan R., Wong H.K. Curve progression in idiopathic scoliosis: follow-up study to skeletal maturity. Spine (Phila Pa 1976) 2009;34(7):697–700. doi: 10.1097/BRS.0b013e31819c9431. [DOI] [PubMed] [Google Scholar]
- 20.Lee C.F., Fong D.Y., Cheung K.M., Cheng J.C., Ng B.K., Lam T.P. A new risk classification rule for curve progression in adolescent idiopathic scoliosis. Spine J. 2012;12(11):989–995. doi: 10.1016/j.spinee.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Collis D.K., Ponseti I.V. Long-term follow-up of patients with idiopathic scoliosis not treated surgically. J. Bone Joint Surg. Am. 1969;51(3):425–445. [PubMed] [Google Scholar]
- 22.Weinstein S.L., Ponseti I.V. Curve progression in idiopathic scoliosis. J. Bone Joint Surg. Am. 1983;65(4):447–455. [PubMed] [Google Scholar]
- 23.Weinstein S.L., Zavala D.C., Ponseti I.V. Idiopathic scoliosis: long-term follow-up and prognosis in untreated patients. J. Bone Joint Surg. Am. 1981;63(5):702–712. [PubMed] [Google Scholar]
- 24.Weinstein S.L., Dolan L.A., Spratt K.F., Peterson K.K., Spoonamore M.J., Ponseti I.V. Health and function of patients with untreated idiopathic scoliosis: a 50-year natural history study. JAMA. 2003;289(5):559–567. doi: 10.1001/jama.289.5.559. [DOI] [PubMed] [Google Scholar]
- 25.Peterson L.E., Nachemson A.L. Prediction of progression of the curve in girls who have adolescent idiopathic scoliosis of moderate severity. Logistic regression analysis based on data from The Brace Study of the Scoliosis Research Society. J. Bone Joint Surg. Am. 1995;77(6):823–827. doi: 10.2106/00004623-199506000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Noshchenko A., Hoffecker L., Lindley E.M., Burger E.L., Cain C.M., Patel V.V. Predictors of spine deformity progression in adolescent idiopathic scoliosis: A systematic review with meta-analysis. World J Orthop. 2015;6(7):537–558. doi: 10.5312/wjo.v6.i7.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogura Y., Kou I., Takahashi Y., Takeda K., Minami S., Kawakami N. A functional variant in MIR4300HG, the host gene of microRNA MIR4300 is associated with progression of adolescent idiopathic scoliosis. Hum. Mol. Genet. 2017;26(20):4086–4092. doi: 10.1093/hmg/ddx291. [DOI] [PubMed] [Google Scholar]
- 28.Mao S.H., Qian B.P., Shi B., Zhu Z.Z., Qiu Y. Quantitative evaluation of the relationship between COMP promoter methylation and the susceptibility and curve progression of adolescent idiopathic scoliosis. Eur. Spine J. 2018;27(2):272–277. doi: 10.1007/s00586-017-5309-y. [DOI] [PubMed] [Google Scholar]
- 29.Baranzini S.E., Mudge J., van Velkinburgh J.C., Khankhanian P., Khrebtukova I., Miller N.A. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464(7293):1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ollikainen M., Ismail K., Gervin K., Kyllonen A., Hakkarainen A., Lundbom J. Genome-wide blood DNA methylation alterations at regulatory elements and heterochromatic regions in monozygotic twins discordant for obesity and liver fat. Clin. Epigenetics. 2015;7:39. doi: 10.1186/s13148-015-0073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roos L., van Dongen J., Bell C.G., Burri A., Deloukas P., Boomsma D.I. Integrative DNA methylome analysis of pan-cancer biomarkers in cancer discordant monozygotic twin-pairs. Clin. Epigenetics. 2016;8:7. doi: 10.1186/s13148-016-0172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong C.C., Meaburn E.L., Ronald A., Price T.S., Jeffries A.R., Schalkwyk L.C. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol. Psychiatry. 2014;19(4):495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaiyasap P., Kulawonganunchai S., Srichomthong C., Tongsima S., Suphapeetiporn K., Shotelersuk V. Whole genome and exome sequencing of monozygotic twins with trisomy 21, discordant for a congenital heart defect and epilepsy. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faulk C., Dolinoy D.C. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics. 2011;6(7):791–797. doi: 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida M., Itano N., Yamada Y., Kimata K. In vitro synthesis of hyaluronan by a single protein derived from mouse HAS1 gene and characterization of amino acid residues essential for the activity. J. Biol. Chem. 2000;275(1):497–506. doi: 10.1074/jbc.275.1.497. [DOI] [PubMed] [Google Scholar]
- 36.Inkinen R.I., Lammi M.J., Agren U., Tammi R., Puustjarvi K., Tammi M.I. Hyaluronan distribution in the human and canine intervertebral disc and cartilage endplate. Histochem. J. 1999;31(9):579–587. doi: 10.1023/a:1003898923823. [DOI] [PubMed] [Google Scholar]
- 37.Roughley P.J., Lamplugh L., Lee E.R., Matsumoto K., Yamaguchi Y. The role of hyaluronan produced by Has2 gene expression in development of the spine. Spine (Phila Pa 1976) 2011;36(14):E914–E920. doi: 10.1097/BRS.0b013e3181f1e84f. [DOI] [PubMed] [Google Scholar]
- 38.Cheng J.C., Qin L., Cheung C.S., Sher A.H., Lee K.M., Ng S.W. Generalized low areal and volumetric bone mineral density in adolescent idiopathic scoliosis. J. Bone Miner. Res. 2000;15(8):1587–1595. doi: 10.1359/jbmr.2000.15.8.1587. [DOI] [PubMed] [Google Scholar]
- 39.Little J.P., Pearcy M.J., Izatt M.T., Boom K., Labrom R.D., Askin G.N. Understanding how axial loads on the spine influence segmental biomechanics for idiopathic scoliosis patients: A magnetic resonance imaging study. Clin Biomech (Bristol, Avon) 2016;32:220–228. doi: 10.1016/j.clinbiomech.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Dickson R.A. The aetiology of spinal deformities. Lancet. 1988;1(8595):1151–1155. doi: 10.1016/s0140-6736(88)91963-0. [DOI] [PubMed] [Google Scholar]
- 41.Newton Ede M.M., Jones S.W. Adolescent idiopathic scoliosis: evidence for intrinsic factors driving aetiology and progression. Int. Orthop. 2016;40(10):2075–2080. doi: 10.1007/s00264-016-3132-4. [DOI] [PubMed] [Google Scholar]
- 42.Cheung K.M., Wang T., Qiu G.X., Luk K.D. Recent advances in the aetiology of adolescent idiopathic scoliosis. Int. Orthop. 2008;32(6):729–734. doi: 10.1007/s00264-007-0393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lonstein J.E., Carlson J.M. The prediction of curve progression in untreated idiopathic scoliosis during growth. J. Bone Joint Surg. Am. 1984;66(7):1061–1071. [PubMed] [Google Scholar]
- 44.Lara T, Astur N, Jones TL, Perake V, Moisan A, Warner WC, Jr., et al. The Risk of Curve Progression and Surgery in African Americans With Adolescent Idiopathic Scoliosis. Spine Deform 2017;5(4):250–4. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5