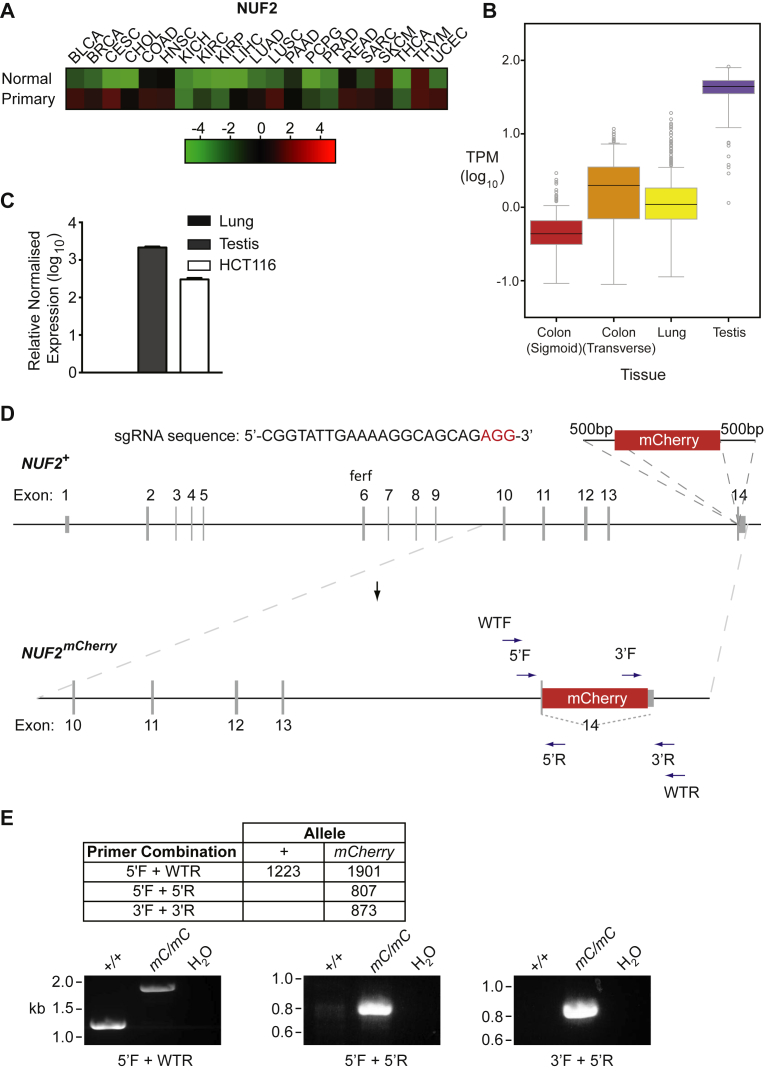
Fig. 1.
Generation of NUF2mCherry/mCherry reporter cell line in HCT116 cells. (A) Heat map of median NUF2 expression scores in normal and primary tumour tissues in the following different cancers: breast invasive carcinoma (BRCA); bladder urothelial carcinoma (BLCA); cholangiocarcinoma (CHOL); colon adenocarcinoma (COAD); head & neck squamous cell carcinoma (HNSC); kidney chromophobe (KICH); kidney renal clear cell carcinoma (KIRC); kidney renal papillary cell carcinoma (KIRP); liver hepatocellular carcinoma (LIHC); lung adenocarcinoma (LUAD); lung squamous cell carcinoma (LUSC); mesothelioma (MESO); prostate adenocarcinoma (PRAD); rectum adenocarcinoma (READ); thyroid carcinoma (THCA); uterine corpus endometrioid carcinoma (UCEC); (B). Box plots showing RNA-seq data of NUF2 expression in different tissues indicated. Log10 expression level measured as Transcripts Per Million (TPM). Lines indicate median and 25th and 75th percentiles. Values shown for each tissue are median and sample size, respectively: Colon (sigmoid) (0.386, 233); Colon (transverse) (1.933, 274); Lung (1.045, 427) and Testis (44.18, 259). Data obtained from the GTex Portal (https://www.gtexportal.org/home/); (C) Bar graphs showing relative, GAPDH-normalised qRT-PCR expression levels of NUF2 in HCT116 cells. Log10 expression levels in human lung and testis are shown as controls (negative and positive, respectively). Expression is shown relative to levels in lung; (D) Diagram showing CRISPR/Cas gene editing strategy used. Sequence of sgRNA used to target hCas9 to the NUF2 locus is shown. Primers used in targeting the NUF2 locus are located outside the homology arms; (E) PCR results of targeted NUF2mCherry/mCherry clones (labelled mC/mC). Table shows expected amplicon sizes using primers indicated in D. Images of gel resolved amplicons using the primer combinations indicated stained with ethidium bromide and visualised under UV light.