Abstract
Background
Neurofibromatosis type I (NF1) is caused by heterozygous loss-of-function variants in the NF1 gene encoding neurofibromin which serves as a tumor suppressor that inhibits RAS signaling and regulates cell proliferation and differentiation. While, the only well-established functional domain in the NF1 protein is the GAP-related domain (GRD), most of the identified non-truncating disease-causing variants are located outside of this domain, supporting the existence of other important disease-associated domains. Identifying these domains may reveal novel functions of NF1.
Methods
By implementing inferential statistics combined with machine-learning methods, we developed a novel NF1-specific functional prediction model that focuses on nonsynonymous single nucleotide variants (SNVs). The model enables annotating all possible NF1 nonsynonymous variants, thus mapping the range of pathogenic non-truncating variants at the codon level across the NF1 gene.
Findings
The generated model demonstrates high absolute prediction value for missense and splice-site variations (area under the ROC curve of 0.96) outperforming 14 other established models.
By reviewing the entire dataset of nonsynonymous variants, two novel domains (Armadillo type fold 1 and 2) were identified as being associated with pathogenicity (OR 1.86; CI 1.04 to 3.34 and OR 2.08; CI 1.08 to 4.04, respectively; P < .05). Specific exons and codons associated with increased pathogenicity were also detected along the gene inside and outside the GRD domain.
Interpretation
The developed model, enabled better prediction of pathogenicity for variants in NF1 gene, as well as elucidation of novel NF1-associated domains in addition to the GRD.
Fund
This work was partially supported by the Kahn foundation. DGE is supported by the all Manchester NIHR Biomedical Research Centre (IS-brC-1215-20007).
Keywords: Machine learning, Neurofibromatosis 1, Functional annotation, Genetic variant, Variant prioritization
Abbreviations: AUC, Area Under the Curve; CALM, Café Au Lait Macules; CI, Confidence Intervals; DRF, Distributed Random Forest; ExAC, Exome Aggregation Consortium; FPR, False Positive Rate; GBM, Gradient Boosting Machine; GLM, Generalized Linera Model; gnomAD, Genome Aggregation Database; GRD, GAP Related Domain; LOVD, Leiden Open Variation Database; mRNA, Messanger Ribonucleic acid; NF1, Neurofibromatosis Type 1; OR, Odds Ratio; ROC, Receiver Operating Characteristic; SNV, Single Nucleotide Variant; UTR, Untranslated Regions; XRT, eXtremely Randomized Trees
Research in context.
Evidence before this study
The American College of Genetics and Genomics recommends that laboratories performing clinical sequencing seek and report disease-causing variants in multiple functional genes including NF1. Missense variants represent a major diagnostic issue in NF1. Although many tools have been developed for prediction of variant pathogenicity, most were developed and trained on the entire genome. We hypothesize that this pan-genome approach may miss important gene-specific attributes contributing to variant functionality.
Added value of this study
Here we describe an exhaustive review of NF1 features associated with increased likelihood of variant functionality. We implement advanced machine learning algorithms in order to develop an NF1-specific variant pathogenicity prediction tool which out-performed all other scoring methods. Using the model, a score was calculated for the entire spectrum of nonsynonymous variants across the gene.
Implications of all the available evidence
This high resolution prediction of pathogenicity allowed us to identify NF1 regions, never before described that may represent novel NF1-therapy targets. This work demonstrates the benefit of gene-centered analysis and may be applied to other functionally important genes.
Alt-text: Unlabelled Box
1. Background
Neurofibromatosis type 1 (NF1, MIM #162200) is an autosomal dominant neurocutaneous disorder with a birth incidence of 1 in 2-2500 and disease prevalence of around 1 in 4000.
[13,49]. The disease is characterized by multiple café au lait macules (CALM), skin-fold freckling, iris Lisch nodules and neurofibromas. In addition, there are a large number of disease complications, which can affect any body system [15]. The disorder is caused by heterozygous loss-of-function variants in the cytoplasmic protein neurofibromin. The protein serves as a tumor suppressor that inhibits RAS signaling and regulates cell proliferation and differentiation. The disease is caused by loss-of function variants, resulting in an increased RAS activity. This increase in RAS activity accounts for both tumorigenesis and neuronal dysfunction (such as learning disabilities, attention deficits) by different mechanisms (for review: [18,21,30]). Given the large size of the gene and the protein (~327 kDa), and the fact that about 50% of cases are caused by de novo variants, multiple disease causing variants have been reported in the public domains. Most detected disease-causing variants are stop-gain (truncating) and start-loss single nucleotide variants (SNVs) and frameshift insertions and deletions which result in protein truncation and a loss-of-function [37]. These variants generate premature stop codons and are expected to lead to mRNA nonsense-mediated decay and decreased protein level. Although some missense and small in-frame variants may result in protein instability or alter splicing and therefore result in decreased protein levels, most act based on their functional effect at the protein level. While overall, missense variants account for <18% of pathogenic variants, they still represent a major diagnostic issue in NF1 [14,37].
So far, the only well-established functional domain in the NF1 protein is the RAS-GAP domain (also known as the GAP-related domain (GRD)) encoded by exons (27–34). However, most of the non-truncating disease-causing variants detected are located outside this domain suggesting the existence of other important domains in the protein that are associated with protein loss-of-function either directly or by disrupting the RAS-GAP domain [36]. The NF1 gene has one of the highest mutation rates and therefore novel variants are likely to be identified as incidental findings in unrelated sequencing studies. As the American College of Genetics and Genomics recommends that laboratories performing clinical sequencing seek and report disease-causing variants in NF1 gene along with 59 other genes related to actionable disorders [26], categorizing non-truncating variants as either pathogenic or benign has important clinical implications.
Here we describe an exhaustive characterization of the entire nonsynonymous variant spectrum in NF1. By implementing both inferential statistical methods and machine learning methods, we examined which genetic features are associated with variant pathogenicity. We developed an NF1-specific functional prediction model that focuses on nonsynonymous SNVs and compared its performance against other established tools. We then use our prediction method to score all possible NF1 nonsynonymous variants, thus mapping the range of non-truncating variants predicted to be pathogenic across the NF1 gene. This extensive high resolution prediction of pathogenicity enabled the elucidation of functional domains across the gene's entire length. We demonstrate the utility of our method through examination of the interaction regions between NF1 and SPRED1. SPRED1 (Sprouty-related protein with an EVH1 domain) gene is a putative tumor suppressor and an important interacting gene with NF1. Much like NF1, SPRED1 is a negative regulator of Ras/MAPK signaling and acts by binding to neurofibromin inducing it's localization in the plasma membrane, which subsequently down-regulates Ras-GTP levels [46]. Recent works characterizing the association between NF1 and SPRED1 uncovered specific essential regions in the N and C terminal regions of the RAS-GAP domain that are required for their interaction [11,22]. An in-depth review of the predicted scores distribution in the N and C terminal regions identified a higher rate of pathogenic variants occurring within these regions and enabled high-resolution assessment of their limits down to the codon level.
We provide a webserver describing the entire set of nonsynonymous and splice site SNVs in NF1 allowing the research and clinical community to explore various genetic annotations including variant type, genetic region, population based allele frequencies, functional prediction scores and our own model's final score (https://isakovlab.shinyapps.io/NF1-VariantAnnotationServer/).
2. Methods
2.1. NF1 gene variant spectrum
The complete spectrum of NF1 SNVs includes 848,250 different variants. We first annotated each variant by its pathogenicity status as either pathogenic, benign or unknown. Pathogenic variants were collected from the Leiden Open Variation Database (LOVD) database [17]. Pathogenicity status is based on the LOVD version used to train the model (05/2017). We note that pathogenicity status may change over time and therefore the annotations used at the time of model development should not be considered as the most up-to-date version. In order to determine which variants are benign we considered each variant's allele frequency in three different databases of healthy populations: The 1000 Genomes Project, The Exome Aggregation Consortium (ExAC) and the Genome Aggregation Database (gnomAD) [33,43]. Since NF1is an autosomal dominant disorder, any variant with an allele frequency above 0 in any of these populations, and that was not previously marked as pathogenic was considered to be benign. Variants that do not fall into either category, were considered as unknown.
2.2. Variant annotations
Known NF1 pathogenic variants were downloaded from LOVD [17]. Only nonsynonymous and synonymous SNVs with a status of pathogenic or probably pathogenic were collected. Variant annotation was performed using SnpEff [6] and included annotations gathered in dbNSFP [35]. All variant and feature positions are reported using the ENST00000356175 Ensembl transcript (NM_000267). Alternatively spliced exons were not included in the analysis as the LOVD variants were specified only in accordance to ENST00000356175. The SnpEff tools allows for functional annotation of each variant (e.g whether it is synonymous/nonsynonymous/stop-gain/etc.) The dbNSFP database, adds various annotations to all potential nonsynonymous SNVs and splice site SNVs, including prediction scores from 16 prediction algorithms (SIFT [38], Polyphen2 [1], LRT [5], MutationTaster [42], MutationAssessor [41], FATHMM [44], MetaSVM, MetaLR [10], CADD [27], VEST [2], PROVEAN [4], M-CAP [24], MutPred [34], Eigen and Eigen PC [23] and dbscSNV [25]) and eight conservation scores (phyloP46way_primate, phyloP46way_placental, phyloP100way_vertebrate [40], phastCons46way_primate, phastCons46way_placental, phastCons100way_veterbrate [45], GERP++ [8] and SiPhy [19]). Healthy population databases included the 1000 genomes project [47], the Exome Aggregation Consortium (ExAC) and the Genome Aggregation Consortium (gnomAD) [33]. NF1 protein domain annotation was performed using Interpro [16] and Nextprot [20].
2.3. Functional prediction model generation
The purpose of the prediction model is to predict whether a given nonsynonymous or splice site variant is pathogenic or not. In order to generate the model we collected only variants that fall into one of these gene effect types (in accordance with sequence ontology terms [12]): missense variant (N = 17,909), missense variant splice region variant (missense variant within a splice site, within 1–3 bases of the exon; N = 738), splice donor intron variant (A splice variant that changes the 2 base pair region at the 5′ end of an intron; N = 126), splice region intron variant (intron variant within a splice site, within 3–8 bases of the intron; N = 226), splice acceptor intron variant (A splice variant that changes the 2 base region at the 3′ end of an intron; N = 116) and splice region synonymous variant (synonymous variant within a splice site, within 1–3 bases of the exon; N = 30). There are 19,145 such variants across the NF1 gene, out of which, a total of 1463 variants were used to generate the model. 436 (30%) of which were known to be pathogenic and 1027 were observed in healthy populations and therefore deemed benign. The dataset was divided into two sets, one for the purpose of training the model (75%) and the other for validation (25%). Each variant in the dataset had 55 different features (i.e annotations) including: the variant's gene effect, amino acid position, exon number, domain regions and rankscores corresponding to the aforementioned, dbNSFP gathered, functional prediction and conservation scores. Categorical features were split into dummy variables resulting in 184 feature annotations. In order to select the most informative features, fifty random forest models were trained. Each model was trained on a different randomly selected variant set comprising 70% of the data. Feature importance was calculated by each model and overall feature importance was determined by the mean importance score. The best feature subset was identified by evaluating a Gradient Boosting Machine (GBM) model trained on an iteratively growing subset of the highest scoring features. The final feature set was composed of the top 75 features. Using the selected feature set, four machine learning algorithms were evaluated [GBM, Generalized Linera Model (GLM), Distributed Random Forest (DRF) and eXtremely Randomized Trees (XRT)]. Briefly, 100 different models were generated and a stacked ensemble learner of the best performing model from each algorithm type was then generated. All models were trained using 10-fold cross validation to minimize the logarithmic loss which penalizes classifiers that are confident about an incorrect classification. Models were trained and validated using the H2O R package [31].
2.4. Statistical analysis
The effect of variant type on pathogenicity was assessed using the Fisher's exact test. In cases where there was a variant type with only pathogenic or benign variants, and there were >100 variants of that type, odds estimations and their corresponding confidence intervals were approximated by adding one to each cell in the contingency table, penalizing the effect size (and test decision) towards no effect. The effect of gene region on pathogenicity was assessed using multivariate logistic regression, adjusting for variant type. ROC curves were compared using DeLong's test for two correlated ROC curves. In order to identify pathogenic codons, a bootstrap method was implemented, simulating 500,000 random score samples and calculating for each codon the rate of samples with an equal or more number of variants with a score higher than the selected threshold of 0.538 (corresponding to a false positive rate (FPR) of 1%). Comparison of the number of variants predicted to be pathogenic between regions was performed using Fisher's exact test.
2.5. Webserver development
The webserver was developed using the shiny web application framework [3], and published on shinyapps.io, a cloud-service platform used to publish applications developed using shiny.
3. Results
3.1. Dataset
Initially we composed a comprehensive list of all the variants with known pathogenicity status across the NF1 gene. The final list of variants included 22,323 variants, 21,572 (96.6%) of which were benign and 751 (3.36%) pathogenic. There were 20,161 (90.3%) intronic variants, 1885 (8.44%) exonic, and 277 (1.24%) within untranslated regions (UTR). The majority of pathogenic variants were within exons (479; 63.8%).
3.2. Factors associated with pathogenicity
We first tested the effect of each variant type on pathogenicity. As expected nonsense variants were highly associated with pathogenicity, followed by splice site and finally missense variants. Synonymous variant as well as variants inside 3’ UTRs or introns were associated with a lower rate of pathogenicity (Supplementary Fig. 1).
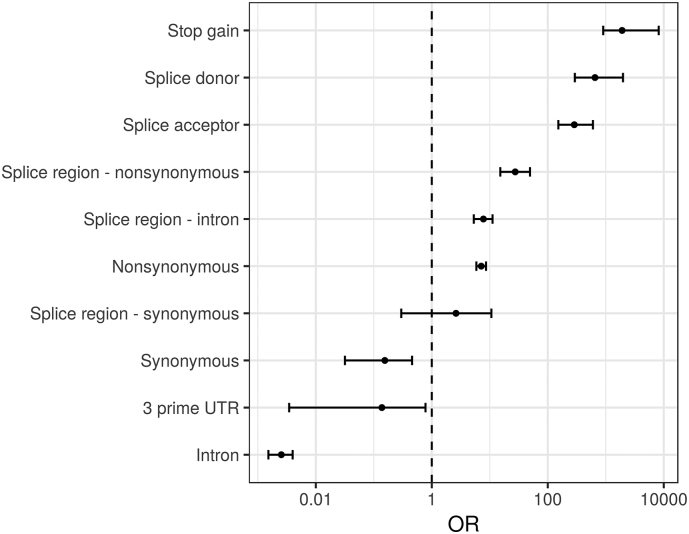
Supplementary Fig. 1.
Genetic effect odds ratio.
Variants were annotated with their predicted genetic effect, the rate of pathogenic variants was calculated for each genetic effect and an odds ratio was calculated in order to rank them according to their association with pathogenicity. Several variant types were significantly associated with pathogenicity: Stop gain (OR 1912.9; 903.92 to 8192; P < 1e-100), splice donor (A splice variant that changes the two base region at the 5′ end of an intron) (OR 651.8; 293 to 1983.1; P < 1e-100), and splice acceptor variants (A splice variant that changes the two base region at the 3′ end of an intron) (OR 286.9; 152.2 to 601.1; P < 1e-100), missense variants in a splice region (within 1–3 bases of the exon) (OR 27.5; 15.2 to 49.4; P < 1e-22), intron variants in a splice region (within 3–8 bases of the intron) (OR 7.8; 5.3 to 11.2; P < 1e-19), and missense variants (OR 7.1; 5.9 to 8.6; P < 1e-68). The following variant types were identified to be associated with a lower rate of pathogenicity: synonymous variants (OR 0.155; 0.032 to 0.457; P < 1e-04) and variants found in the 3 prime UTR (OR 0.138; 0.003 to 0.779; P < 0.01) and inside introns (OR 0.003; 0.002 to 0.004; P < 1e-100).
We extended the search for factors that contribute to variant pathogenicity with genetic domains found within the NF1 gene. A multivariate logistic regression was used in order to examine the association of each feature with pathogenicity after adjusting for variant type. For this analysis we did not consider variant types that invariably result in loss-of-function (stop gain and start loss) or have no recorded pathogenic variant (UTR variants). In accordance to previous studies, we demonstrated the RAS GTPase activation protein domain is associated with pathogenicity (OR 3.24; 1.14 to 9.18; P < 0.05). Additionally, the Armadillo type fold 1, which extends for almost the entire gene (exons 8–54) and Armadillo type fold 2 (exons 37–48) domains presented a significant association with pathogenicity (OR 1.86; CI 1.04 to 3.34 and OR 2.08; CI 1.08 to 4.04, respectively; P < 0.05).
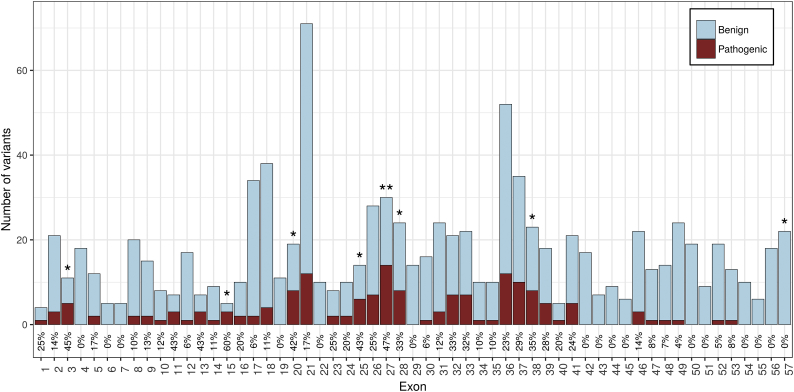
Next we examined whether exons across the NF1 gene have different effect on pathogenicity. After correcting for multiple hypothesis testing, the only exon with a significantly higher proportion of pathogenic variants was exon 27 (OR 4.63; 2.05 to 10.37; P < 0.05, Supplementary Fig. 2.
Supplementary Fig. 2.
Per-exon pathogenicity rate.
In order to identify exons across the NF1 gene that have different effects on pathogenicity, we compared each exon's pathogenic variants rate against all the rest. Although initially 8 exons were deemed significant (*) only one remained after correcting for multiple hypothesis testing (**).
In order to increase the analysis resolution, we reviewed all the exons that were deemed significantly associated with pathogenicity before multiple hypothesis correction (exons 3, 15, 20, 25, 27, 28, 38 and 57, Supplementary Fig. 2). Each exon was split into several parts with approximately 7 variants in each part, resulting in the minimum number of variants required in order to identify a strong association. This analysis pinpointed the following codons as having more than ten times the likelihood of carrying a pathogenic variant than the rest of the coding sequence (Table 1): codons 777–793 in exon 20 (OR 10.23; CI 2.7–47.01; P < 0.005), codons 1209–1222 in exon 27 (OR 10.58; CI 2.23–66.18; P < 0.01) and codons 1082–1104 in exon 25 (OR 12.67; CI 2.05–134.36; P < 0.05).
Table 1.
NF1 codons with a high rate of pathogenic variants.
Exons with a significantly higher rate of pathogenic variants were identified and selected for subsequent in-depth codon analysis (exons 3, 15, 20, 25, 27, 28, 38 and 57). Each exon was split according to the minimal number of variants required in order to identify a strong association. This table describes the codons having more than ten times the likelihood of carrying a pathogenic variant than the rest of the coding sequence.
| Exon | Codons | Benign | Pathogenic | Or | Conf.Low | Conf.High | P.Value | Corrected.P |
|---|---|---|---|---|---|---|---|---|
| 20 | [777,793) | 4 | 8 | 10.232 | 2.702 | 47.012 | 0.000 | 0.003 |
| 20 | [793,802] | 7 | 0 | 0.000 | 0.000 | 3.602 | 0.606 | 0.664 |
| 25 | [1068,1082) | 6 | 1 | 0.848 | 0.018 | 7.066 | 1.000 | 1.000 |
| 25 | [1082,1104] | 2 | 5 | 12.670 | 2.052 | 134.364 | 0.002 | 0.011 |
| 27 | [1175,1192) | 4 | 4 | 5.295 | 0.975 | 28.772 | 0.027 | 0.091 |
| 27 | [1192,1209) | 3 | 4 | 7.055 | 1.181 | 48.588 | 0.015 | 0.066 |
| 27 | [1209,1222) | 3 | 6 | 10.574 | 2.229 | 66.183 | 0.001 | 0.008 |
| 27 | [1222,1234] | 6 | 0 | 0.000 | 0.000 | 4.556 | 0.598 | 0.664 |
| 28 | [1241,1255) | 6 | 2 | 1.700 | 0.166 | 9.621 | 0.625 | 0.664 |
| 28 | [1255,1272) | 5 | 3 | 3.057 | 0.470 | 15.894 | 0.132 | 0.249 |
| 28 | [1272,1284] | 5 | 3 | 3.057 | 0.470 | 15.894 | 0.132 | 0.249 |
| 38 | [1861,1883) | 5 | 3 | 3.061 | 0.471 | 15.914 | 0.132 | 0.249 |
| 38 | [1883,1896) | 5 | 3 | 3.061 | 0.471 | 15.914 | 0.132 | 0.249 |
| 38 | [1896,1909] | 5 | 2 | 2.042 | 0.193 | 12.611 | 0.324 | 0.516 |
| 57 | [2774,2787) | 8 | 0 | 0.000 | 0.000 | 2.852 | 0.364 | 0.516 |
| 57 | [2787,2808) | 8 | 0 | 0.000 | 0.000 | 2.852 | 0.364 | 0.516 |
| 57 | [2808,2818] | 6 | 0 | 0.000 | 0.000 | 4.131 | 0.597 | 0.664 |
3.3. Functional and conservation predictions
Functional and conservation prediction tools assist in the task of causative variant prioritization and identification. Most employ advanced statistical methods such as logistic regression, hidden markov models, random forest, support vector machines and neural networks to derive functionality from a pre-compiled list of genetic annotations including sequence homology, mRNA and regulatory features, secondary and tertiary structure, conservation, epigenomic signals and multiple sequence alignment. During development, these tools were trained on lists of known Mendelian disease associated variations and their performance evaluated on the entire data set of variations. Since these tools weigh in many factors and are specifically trained to identify variants predicted to be pathogenic, reviewing the predicted score for each individual variant might help pinpoint variants with high probability for pathogenicity. Moreover, regions with high overall scores may be uncovered and provide additional insight regarding how different regions are associated with pathogenicity. We hypothesized that since most functional prediction tools were trained on pathogenic variants associated with a multitude of Mendelian diseases, we might be able to improve overall prediction performance by training a new model specifically on NF1 gene variants.
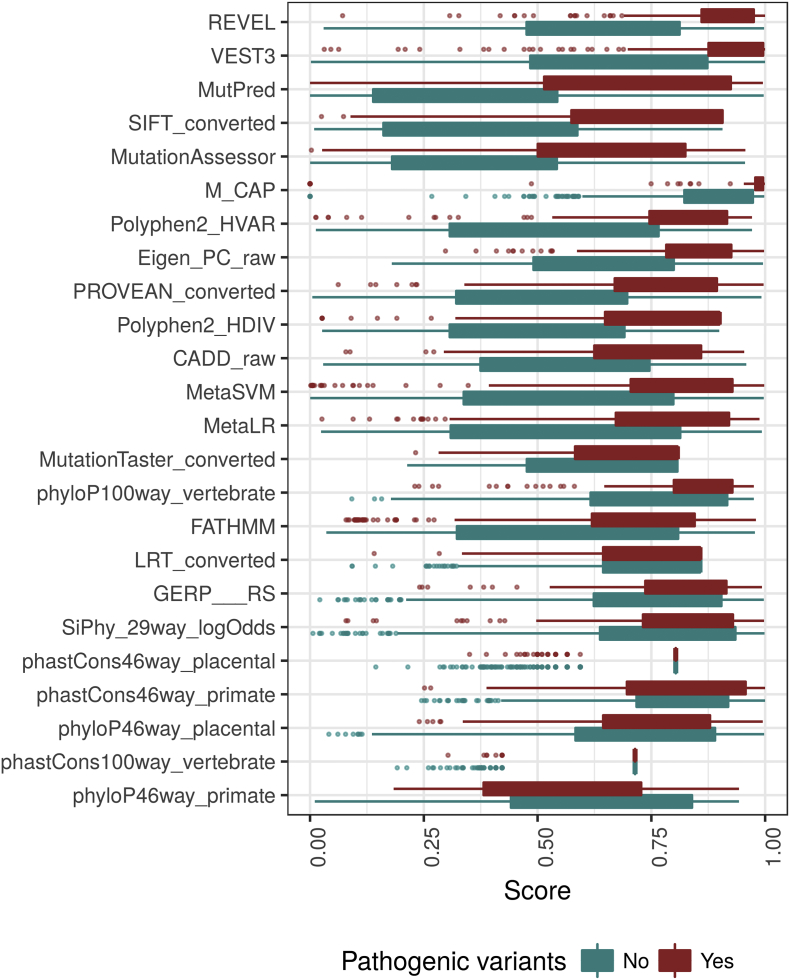
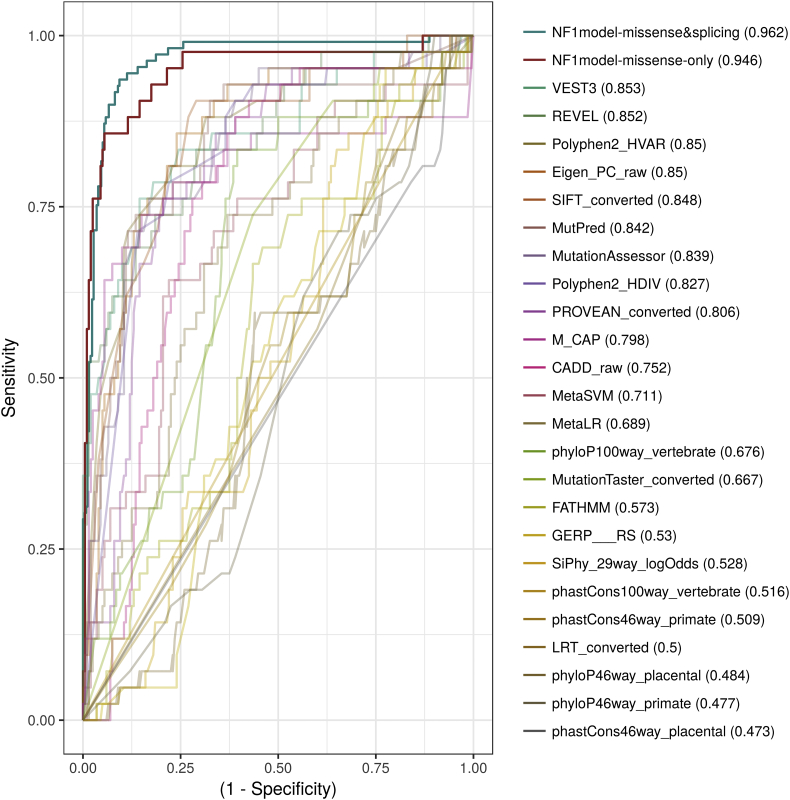
In order to generate such a model, we first explored how established prediction tools perform on known NF1 pathogenic variations. Initially, for each tool, we assessed the magnitude of the difference between the scores of pathogenic variants and those of non-pathogenic variants. The top five tools with the highest median score difference (Mann-Whitney test; P < 1e-30) were REVEL (0.24; CI 0.21–0.27), VEST3 (0.19 CI 0.16–0.22), MutPred (0.37 CI 0.28–0.32), SIFT (0.32 CI 0.28–0.36) and MutationAssessor (0.3 CI 0.26–0.34) (Supplementary Fig. 3). We continued and compared each tool's performance under various threshold settings by plotting the receiver operating characteristic (ROC) curve, and overall performance by calculating the area under the curve (AUC). The top 5 tools with the highest AUC were VEST3 (0.853), REVEL (0.852), Polyphen2-HVAR (0.85), Eigen-PC (0.85) and SIFT (0.848) (Fig. 1).
Supplementary Fig. 3.
Prediction tool performance.
Each functional prediction tool produces a score for each variant, we compared the scores given by each tool to the known pathogenic and benign variants. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles), the whiskers correspond to 1.5* the inter quantile range (IQR) and the points are outliers. If the difference is large, the tool is considered to perform well in the task of differentiating benign from pathogenic variants. The tools at the top demonstrated the best performance.
Fig. 1.
Model performance.
In order to compare the performance of the NF1-specific model and the other available functional prediction tools, we used a test variants dataset that was not used during the training of the model. These variants were scored by each of the tools and the model and the area under the receiver operating characteristic curve was calculated. The NF1-specific model demonstrated significant improvement in performance on the test set which includes only nonsynonymous variants and even better when including variants effecting splice sites as well.
In order to train our own prediction model, we used as input the scores provided by each prediction tool, together with various other annotations including: the exon number, the amino acid position, the genetic type of variant (splice site, nonsynonymous etc.) and which genetic domains the variant affects. The model was trained to predict how likely a variant is to be pathogenic using a training set that includes 1097 nonsynonymous and splice site variants, 327 (29.9%) of which are known to be pathogenic. The final model was based on a stacked ensemble of four different machine learning algorithms (See details in methods section). We compared the performance of the generated model against the other tools on a test set of nonsynonymous variants which were not used during the training of the model (Fig. 1). The model demonstrated a significantly higher AUC value than the tool with the highest AUC (0.946; P < 0.05). Since most tools focus on predicting the effect of nonsynonymous variants, the aforementioned test set did not include splicing variants. Moreover, since the NF1 gene is expressed in peripheral white blood cells and splicing of the NF1 mRNA can be readily studied in a diagnostic lab, in-silico functional prediction of such variants holds less value. However, our model was trained on both types of variants and when testing its performance on a test set that includes both nonsynonymous and splicing variants the model demonstrated an improved AUC of 0.962 (Fig. 1). In order to allow researchers and clinicians to evaluate NF1 variants of interest, we generated a webserver that includes the entire set of NF1 nonsynonymous and splice site variants with various annotations, including our model's final score (https://isakovlab.shinyapps.io/NF1-VariantAnnotationServer/).
4. Discussion
4.1. Coding effect and pathogenicity
Here we present an exhaustive study which brings to light the genetic factors associated with pathogenicity in the NF1 gene. We begin by reviewing the different types of variants according to their coding effect. Each variant type was compared against all other types in order to identify its association with pathogenicity. Expectedly, synonymous variants were significantly less likely to be pathogenic than missense variants (OR 0.027, CI 0.006 to 0.082; P < 10−10). Stop gain (i.e nonsense) variants, which result in protein truncation and loss-of-function due to premature stop codon transcription, were naturally found to be the most pathogenic type of variant. Out of 274 stop gain variants included in the study, 268 (97.8%) were known to be pathogenic. The remaining 6 had an extremely low allele frequency (<10−6), which is significantly less than the allele frequency of non-pathogenic missense variants (P < 0.03) raising the possibility that these variants might have a milder phenotype that has been misclassified as normal. Variant types with slightly lower pathogenicity rate than stop-gain, were splice donor and acceptor variants (within two base pairs from the 5 and 3 prime ends of the intron, respectively). A previous meta-analysis of 478 disease-associated splicing mutations, in 38 different genes suggested a significantly higher rate of disease-causing variants within the splice donor sites when compared to the splice acceptor site [32]. Our study shows that such a difference is not found in the NF1 gene. Although splice donor variants demonstrated a slightly higher pathogenicity rate than acceptor variants, the difference between these two types of variants was not significant (P = 0.098). Missense variants occurring within the first 1–3 bases of an exon were identified as more likely to be pathogenic than missense variants occurring downstream (OR 4.57; CI 2.48 to 8.42; P < 10−6).
4.2. Genetic domain and pathogenicity
Although the genetic architecture of the NF1 gene has been well studied, the importance of the various domains across the gene and how they are associated with pathogenicity remains unclear. In order to identify genetic regions with possible functional significance we perform multivariate analysis including five different genetic regions across the gene (RAS-GAP, Armadillo-type fold 1 and 2, and CRAL-trio and lipid binding domain). Correcting for variant type, 3 regions were shown to be significantly associated with pathogenicity (RAS-GAP and Armadillo-type fold 1 and 2). The RAS-GAP domain (i.e catalytic domain), which represents a genetic region common to all Ras GTPase-activating proteins, has been previously shown to play a critical role in NF1 pathogenesis [28]. The armadillo type folds are superhelical structural domains with an extensive solvent-accessible surface that favors binding of large substrates such as proteins and nucleic acids.
The armadillo type fold 1 domain extends across 46 exons and includes the RAS-GAP and the armadillo type fold 2. However even after adjusting for these domains, it demonstrates an association with pathogenicity. The Armadillo type 2 fold represents a novel domain of interest in the NF1 gene demonstrating a 2 fold increase in the likelihood of being pathogenic.
4.3. Model based architecture investigation
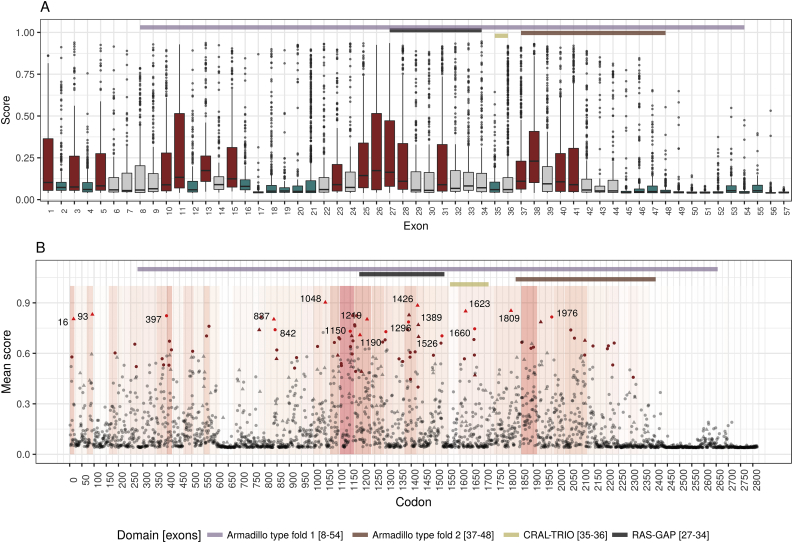
This work describes the development of an NF1-specific pathogenicity prediction model. The model demonstrated improved performance when compared against other established prediction tools. This gene-centered approach may be used on additional genes in the future, in order to facilitate the identification of attributes contributing to pathogenicity that would have otherwise be missed by generating a model based on the entire dataset of genes. Even though scores given by the model were optimized to correspond to the actual probability of pathogenicity, the overall accuracy in the validation set was 92.34%, suggesting the existence of additional phenotype-determining factors or interactions which were not incorporated into the model. Specific therapies for NF1 complications have developed during recent years. These treatments attempt to inhibit the only pathway which is known to be associated with the disease, the RAS-MAPK pathway, which is activated in NF1. Indeed, Selumetinib, a selective inhibitor of MAPK kinase pathway was shown recently to be the first agent inducing a partial responses in NF1–Related Plexiform Neurofibromas [9]. In order to identify additional domains associated with pathogenicity in the NF1 gene, we collected and annotated the entire dataset of known nonsynonymous NF1 variants. Contrary to truncating variants, the pathogenicity of these variants is expected to be influenced by various factors such as their biophysical properties (e.g bulkiness and charge) and their location within the secondary RNA and protein structure. Various regions throughout the gene were compared and those with a significantly higher rate of pathogenic variants were identified. This approach is limited by the relatively low number of known pathogenic variants across the gene. Regions that carry a low number of variants cannot be confidently identified as having any association with pathogenicity. Moreover, this approach cannot be used to study regions that do not have any known variants in them. In order to overcome this challenge, we employed the generated prediction tool to score all possible nonsynonymous variants in the NF1 gene. Since model scores were optimized to correspond to the actual posterior probability of pathogenicity, reviewing the spectrum of scores, results in a high resolution map of pathogenicity across the entire gene (Supplementary Table 1). Initially, an exon-wide analysis of the rate of variants predicted to be pathogenic by the model was performed (Fig. 2A). Two main exonic regions were identified as having a significantly higher rate of pathogenic variants: exons 25–28 and exons 37–41 (with the exception of exon 39). These exons correspond to the 5 prime regions of the RAS-GAP and the Armadillo type fold 2 domains respectively. Consistently, exon 27 had one of the highest rates of pathogenic variants (OR 3.32; CI 2.7 to 4.07; P < 1 × 10−20) with only exons 26 and 38 demonstrating higher rates (OR 4.13 and 3.65 respectively). Although the analysis based exclusively on known-variants suggested there is a lower rate of pathogenic variants in the 3 prime region of the gene, there was not enough information to identify a significant difference. The model based analysis identified a significant decline in pathogenicity rate starting from exon 45 down to the last exon (57), suggesting that nonsynonymous variants found downstream of exon 45 are most likely benign. As of date, this is the first report describing such a reduction in pathogenicity across the 3 prime end of the NF1 gene. Such exon specific functional predictions can be utilized in designing minigenes for in vivo elucidation of regulatory elements and other regulators of pre-mature RNA splicing [7].
Fig. 2.
Model-based analysis.
After generating the NF1-specific prediction model, a score was calculated for the entire dataset of known nonsynonymous NF1 variants. Reviewing the rate of variants predicted to be pathogenic by our model in each exon (A), two main exonic regions were identified as having a significantly higher rate of pathogenic variants (red): exons 25–28 and exons 37–41. These exons correspond to the 5 prime regions of the RAS-GAP domain and the Armadillo type fold 2 domain respectively. The model based analysis also identified a significant decline in pathogenicity rate (blue) starting from exon 45 down to the last exon (57). With pathogenicity scores predicted for every possible nonsynonymous variant, specific codons with higher pathogenicity association could be identified (B). After correction for multiple hypothesis, 85 codons were found to have significantly more variants with a score higher than 0.538 (brown color; corresponding to a FPR of 1%) than would be expected (P < 0.01). In 17 of these codons, all of the variants were above the threshold (red color). While some of these codons already have known pathogenic variants in them (triangle), some represent a novel deleterious loci (point). The background represents the overall exon's odds ratio (with red representing positive association).
Variants within crucial genetic regions are expected to alter gene function. Therefore, codons with a high rate of variants predicted to be pathogenic may serve as markers for such functional loci. Since a pathogenicity score was predicted for every possible nonsynonymous variant, we were now able to identify specific codons predicted to be pathogenic (Fig. 2B). After correction for multiple hypothesis, 85 codons were found to have significantly more variants with a score higher than 0.538 (corresponding to an FPR of 1%) than would be expected (P < 0.05)(Supplementary Table 2). Seventeen codons exclusively harbored variants predicted to be pathogenic (16, 93, 397, 837, 842, 1048, 1150, 1190, 1219, 1296, 1389, 1426, 1526, 1623, 1660, 1809 and 1976). Of these, six were located within the well-known GAP domain and eight (397, 842, 1150, 1296, 1389, 1526, 1660 and 1976) represent novel loci with none of the variants already known to be pathogenic. Our model was trained to predict the probability of pathogenicity and not phenotype severity, therefore the aforementioned codons include both variants associated with a mild form of NF1 such as p.Arg1809Cys [39] and codons adjacent to regions previously associated with a more severe phenotype [29].
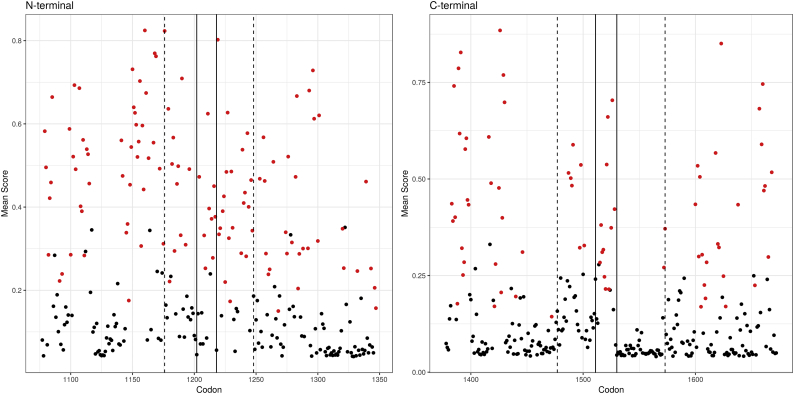
The SPRED1, a putative tumor suppressor and an important interacting gene with NF1, binds to neurofibromin, through an interaction between the EVH1 and the GRD domains. More specifically, EVH1 binds to the GRD boundary regions formed by residues located at the N (Codons 1209–1220) and C (Codons 1477–1573) terminal parts of the GRD domain (With codons 1202–1217 and 1511 and 1530 identified as essential regions [22]). We therefore focused our analysis on the EVH1 domain binding sites. Indeed, the initial codon analysis identified codons 1209–1220 to be associated with pathogenicity (Table 1). Comparing the number of codons with at least one variant predicted to be pathogenic between the EVH1 binding regions and the adjacent regions (100 base pairs on each side) also showed an increase in variants predicted to be pathogenic in the N-terminal binding region (OR 2.19; 1.22 to 3.99; P = 0.005) but only a trend in the C-terminal (OR 2.18; 0.9 to 5.1; P = 0.072) (Fig. 3). Since the model was used to score all possible missense variants across the binding domains, a high-resolution review of the C-terminal region was possible and a drop in predicted pathogenicity rate beyond codon 1528 was demonstrated, suggesting lower binding activity beyond that point, in agreement with Hirata et al.
Fig. 3.
NF1-SPRED1 binding region.
SPRED1 interacts with NF1 by binding to the N and C terminal regions of the RAS-GAP domain. The number of codons with at least one variant predicted to be pathogenic by the model (score above 0.538; corresponding to a FPR of 1%) was compared between these putative binding regions and the adjacent regions (100 base pairs on each side) showed an increase in variants predicted to be pathogenic in the N-terminal binding region (OR 2.19; 1.22 to 3.99; P = 0.005) but only a trend in the C-terminal (OR 2.18; 0.9 to 5.1; P = 0.072). Generating a score across all the codons identified a drop in pathogenicity rate beyond codon 1528. The dashed line marks the borders of the known binding region (codons 1176–1248 and 1477–1573 for the N and C terminal regions, respectively) and the solid line marks the borders of regions previously described as essential (Codons 1202–1217 and 1511 and 1530).
4.4. Model limitations
Even though our model outperformed existing prediction tools, performance was not perfect (Overall accuracy of 92.34% on the validation set), suggesting the existence of additional phenotype-determining factors or interactions which may have been missed during model development. In order to mitigate this uncertainty, the scores generated by the model were optimized to correspond to the overall probability of pathogenicity and were shown to be well calibrated. Currently, there are only a few established NF1 genotype-phenotype associations. Thus, phenotype severity information was not incorporated into model training and model scores correlate only to the variant's probability of resulting in an NF1 phenotype and have no predictive value in regards to overall phenotype severity. Model training was based on the fact that NF1 has an autosomal dominant mode of inheritance with complete penetrance. Therefore any variant with a population frequency above 0 was considered to be benign. However, NF1 has extreme phenotype variability and therefore undiagnosed patients with a milder form may have been incorporated into the healthy population variant databases. Although such misclassification of pathogenic variants as benign during training might lead to biased estimations, their overall frequency is expected to be low and therefore their relative impact on overall model performance is expected to be minor. Finally, we note that although gene-specific in-silico analysis such as the one described here may facilitate variant prioritization and functional assessment, functional characterization remains the gold standard for the definitive classification of variants [48,50].
5. Conclusion
This study presents an exhaustive analysis of the neurofirbomin gene variant spectrum. Inferential statistics were used to identify regions with a high rate of pathogenic nonsynonymous variants likely corresponding to important functional domains. A novel supervised machine learning algorithm was subsequently trained to differentiate functional from benign variants. The developed NF1-specific model outperformed other established prediction scores. Scoring the entire spectrum of nonsynonymous variants across the gene we characterize likely pathogenic regions with unprecedented resolution down to the specific codon level. We believe that this new data may facilitate both improved characterization of putative domains and the detection of novel domains in neurofibromin. The identification of such novel drugable domains may eventually lead to improved treatment and patient care.
The following are the supplementary data related to this article.
Annotated nonsynonymous and splice site NF1 variants.
The generated NF1-specific prediction model was used to score all possible nonsynonymous and splice site variants in the NF1 gene, excluding nonsense variants. The final variant list includes 19,145 variants with various genetic annotations including variant type, genetic region, population based allele frequencies, functional prediction scores and our own model's final score. We note that the known pathogenicity status is based on the LOVD version used to train the model (05/2017) and should not be used as an up-to-date annotation. In order to facilitate search and use of the variant table, we also provide a public webserver with all the annotated variants (https://isakovlab.shinyapps.io/NF1-VariantAnnotationServer/).
Selected codons with high predicted pathogenicity.
Using the NF1-specific model, a pathogenicity score was predicted for every possible nonsynonymous variant. Using these scores codons with a high rate of variants predicted to be pathogenic were identified. After correction for multiple hypothesis, 85 codons were found to have significantly more variants with a score higher than 0.538 (corresponding to an FPR of 1%) than would be expected (P < 0.05). Seventeen codons exclusively harbored variants predicted to be pathogenic (16, 93, 397, 837, 842, 1048, 1150, 1190, 1219, 1296, 1389, 1426, 1526, 1623, 1660, 1809 and 1976). Of these, six were located within the well-known GAP domain and eight (397, 842, 1150, 1296, 1389, 1526, 1660 and 1976) represent novel loci with none of the variants already known to be pathogenic.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Known NF1 pathogenic variants were downloaded from LOVD: http://grenada.lumc.nl/LSDB_list/lsdbs/NF1
Variant annotation included annotations gathered in dbNSFP: https://sites.google.com/site/jpopgen/dbNSFP
The datasets generated during the current study are available in the supplementary material (Supplementary Table 1) and on an online webserver https://isakovlab.shinyapps.io/NF1-VariantAnnotationServer/
Competing interests
The authors declare that they have no competing interests.
Funding
This work was partially supported by the Kahn foundation. DGE is supported by the all Manchester NIHR Biomedical Research Centre (IS-brC-1215-20007).
Authors' contributions
OI and SBS conceived and designed the study. OI collected, analyzed and interpreted the data described in this study. OI, SBS, DW and GE were involved in drafting the manuscript and revising it critically. All authors read and approved the final manuscript.
Contributor Information
Ofer Isakov, Email: oferis@tlvmc.gov.il.
Deeann Wallis, Email: dwallis@uab.edu.
D. Gareth Evans, Email: Gareth.Evans@mft.nhs.uk.
Shay Ben-Shachar, Email: shayb@tlvmc.gov.il.
References
- 1.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter H., Douville C., Stenson P.D., Cooper D.N., Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14(Suppl. 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang W., Cheng J., Allaire J.J., Xie Y., McPherson J., (RStudio, library) 2018. Shiny: Web Application Framework for R. [Google Scholar]
- 4.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:0–1. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper T.A. Use of minigene systems to dissect alternative splicing elements. Methods San Diego Calif. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dombi E., Baldwin A., Marcus L.J., Fisher M.J., Weiss B., Kim A. Activity of Selumetinib in Neurofibromatosis Type 1-Related Plexiform Neurofibromas. N Engl J Med. 2016;375:2550–2560. doi: 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunzendorfer-Matt T., Mercado E.L., Maly K., McCormick F., Scheffzek K. The neurofibromin recruitment factor Spred1 binds to the GAP related domain without affecting Ras inactivation. Proc Natl Acad Sci. 2016;113:7497–7502. doi: 10.1073/pnas.1607298113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eilbeck K., Lewis S.E., Mungall C.J., Yandell M., Stein L., Durbin R. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans D.g., Howard E., Giblin C., Clancy T., Spencer H., Huson S.m. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet A. 2010;152A:327–332. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- 14.Evans D.G., Bowers N., Burkitt-Wright E., Miles E., Garg S., Scott-Kitching V. Comprehensive RNA analysis of the NF1 gene in classically affected NF1 affected individuals meeting NIH criteria has high sensitivity and mutation negative testing is reassuring in isolated cases with pigmentary features only. EBioMedicine. 2016;7:212–220. doi: 10.1016/j.ebiom.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferner R.E., Huson S.M., Thomas N., Moss C., Willshaw H., Evans D.G. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet. 2007;44:81–88. doi: 10.1136/jmg.2006.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fokkema I.F.A.C., Taschner P.E.M., Schaafsma G.C.P., Celli J., Laros J.F.J., den Dunnen J.T. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 18.Friedman J.M. Neurofibromatosis 1. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J., Mefford H.C., Stephens K., Amemiya A., Ledbetter N., editors. GeneReviews(®) University of Washington; Seattle, Seattle (WA): 1993. [Google Scholar]
- 19.Garber M., Guttman M., Clamp M., Zody M.C., Friedman N., Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudet P., Michel P.-A., Zahn-Zabal M., Britan A., Cusin I., Domagalski M. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 2017;45:D177–D182. doi: 10.1093/nar/gkw1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutmann D.H., Parada L.F., Silva A.J., Ratner N. Neurofibromatosis type 1: modeling CNS dysfunction. J Neurosci Off J Soc Neurosci. 2012;32:14087–14093. doi: 10.1523/JNEUROSCI.3242-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirata Y., Brems H., Suzuki M., Kanamori M., Okada M., Morita R. Interaction between a Domain of the negative Regulator of the Ras-ERK Pathway, SPRED1 Protein, and the GTPase-activating Protein-related Domain of Neurofibromin is Implicated in Legius Syndrome and Neurofibromatosis Type 1. J Biol Chem. 2016;291:3124–3134. doi: 10.1074/jbc.M115.703710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [10.1038/ng.3477] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 25.Jian X., Boerwinkle E., Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med Off J Am Coll Med Genet. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 27.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klose A., Ahmadian M.R., Schuelke M., Scheffzek K., Hoffmeyer S., Gewies A. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum Mol Genet. 1998;7:1261–1268. doi: 10.1093/hmg/7.8.1261. [DOI] [PubMed] [Google Scholar]
- 29.Koczkowska M., Chen Y., Callens T., Gomes A., Sharp A., Johnson S. Genotype-phenotype correlation in NF1: evidence for a more Severe phenotype associated with missense mutations affecting NF1 codons 844–848. Am J Hum Genet. 2018;102:69–87. doi: 10.1016/j.ajhg.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krab L.C., Goorden S.M.I., Elgersma Y. Oncogenes on my mind: ERK and MTOR signaling in cognitive diseases. Trends Genet TIG. 2008;24:498–510. doi: 10.1016/j.tig.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Kraljevic T., H2O.ai . 2018. H2O: R Interface for “H2O”. [Google Scholar]
- 32.Krawczak M., Thomas N.S.T., Hundrieser B., Mort M., Wittig M., Hampe J. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 33.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B., Krishnan V.G., Mort M.E., Xin F., Kamati K.K., Cooper D.N. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinforma Oxf Engl. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: a One-Stop Database of Functional predictions and Annotations for Human Non-synonymous and Splice Site SNVs. Hum Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattocks C., Baralle D., Tarpey P., Ffrench-Constant C., Bobrow M., Whittaker J. Automated comparative sequence analysis identifies mutations in 89% of NF1 patients and confirms a mutation cluster in exons 11–17 distinct from the GAP related domain. J Med Genet. 2004;41:e48. doi: 10.1136/jmg.2003.011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Messiaen L.M., Wimmer K. NF1 Mutational Spectrum. Neurofibromatosis. 2008:63–77. [Google Scholar]
- 38.Ng P.C., Henikoff S. SIFT: predicting Amino Acid changes that Affect Protein Function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinna V., Lanari V., Daniele P., Consoli F., Agolini E., Margiotti K. P.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur J Hum Genet EJHG. 2015;23:1068–1071. doi: 10.1038/ejhg.2014.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of Nonneutral Substitution rates on Mammalian Phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr407. gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz, J.M., Rödelsperger, C., Schuelke, M., Seelow, D., n.d. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Metods 7, 575–576. 10.1038/nmeth0810-575. [DOI] [PubMed]
- 43.Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L.A., Edwards K.J. Predicting the functional, molecular, and phenotypic consequences of amno acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K. Evolutionarily Conserved elements in Vertebrate, Insect, Worm, and yeast Genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stowe I.B., Mercado E.L., Stowe T.R., Bell E.L., Oses-Prieto J.A., Hernández H. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev. 2012;26:1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas L., Spurlock G., Eudall C., Thomas N.S., Mort M., Hamby S.E. Exploring the somatic NF1 mutational spectrum associated with NF1 cutaneous neurofibromas. Eur J Hum Genet. 2012;20:411–419. doi: 10.1038/ejhg.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uusitalo E., Leppävirta J., Koffert A., Suominen S., Vahtera J., Vahlberg T. Incidence and mortality of neurofibromatosis: a total population study in Finland. J Invest Dermatol. 2015;135:904–906. doi: 10.1038/jid.2014.465. [DOI] [PubMed] [Google Scholar]
- 50.Wallis D., Li K., Lui H., Hu K., Chen M.-J., Li J. Neurofibromin (NF1) genetic variant structure-function analyses using a full-length mouse cDNA. Hum Mutat. 2018;39:816–821. doi: 10.1002/humu.23421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotated nonsynonymous and splice site NF1 variants.
The generated NF1-specific prediction model was used to score all possible nonsynonymous and splice site variants in the NF1 gene, excluding nonsense variants. The final variant list includes 19,145 variants with various genetic annotations including variant type, genetic region, population based allele frequencies, functional prediction scores and our own model's final score. We note that the known pathogenicity status is based on the LOVD version used to train the model (05/2017) and should not be used as an up-to-date annotation. In order to facilitate search and use of the variant table, we also provide a public webserver with all the annotated variants (https://isakovlab.shinyapps.io/NF1-VariantAnnotationServer/).
Selected codons with high predicted pathogenicity.
Using the NF1-specific model, a pathogenicity score was predicted for every possible nonsynonymous variant. Using these scores codons with a high rate of variants predicted to be pathogenic were identified. After correction for multiple hypothesis, 85 codons were found to have significantly more variants with a score higher than 0.538 (corresponding to an FPR of 1%) than would be expected (P < 0.05). Seventeen codons exclusively harbored variants predicted to be pathogenic (16, 93, 397, 837, 842, 1048, 1150, 1190, 1219, 1296, 1389, 1426, 1526, 1623, 1660, 1809 and 1976). Of these, six were located within the well-known GAP domain and eight (397, 842, 1150, 1296, 1389, 1526, 1660 and 1976) represent novel loci with none of the variants already known to be pathogenic.