Fig. 2.
Model-based analysis.
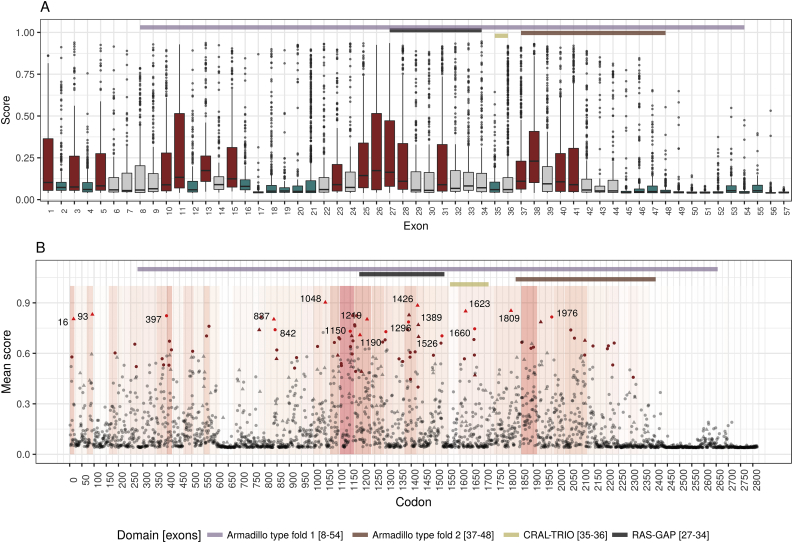
After generating the NF1-specific prediction model, a score was calculated for the entire dataset of known nonsynonymous NF1 variants. Reviewing the rate of variants predicted to be pathogenic by our model in each exon (A), two main exonic regions were identified as having a significantly higher rate of pathogenic variants (red): exons 25–28 and exons 37–41. These exons correspond to the 5 prime regions of the RAS-GAP domain and the Armadillo type fold 2 domain respectively. The model based analysis also identified a significant decline in pathogenicity rate (blue) starting from exon 45 down to the last exon (57). With pathogenicity scores predicted for every possible nonsynonymous variant, specific codons with higher pathogenicity association could be identified (B). After correction for multiple hypothesis, 85 codons were found to have significantly more variants with a score higher than 0.538 (brown color; corresponding to a FPR of 1%) than would be expected (P < 0.01). In 17 of these codons, all of the variants were above the threshold (red color). While some of these codons already have known pathogenic variants in them (triangle), some represent a novel deleterious loci (point). The background represents the overall exon's odds ratio (with red representing positive association).