Abstract
MicroRNAs (miRNAs) are key regulators of different human processes that represent a new promising class of cancer therapeutics or therapeutic targets. Indeed, in several tumor types, including non-small-cell lung carcinoma (NSCLC), the deregulated expression of specific miRNAs has been implicated in cell malignancy. As expression levels of the oncosuppressor miR-34c-3p are decreased in NSCLC compared to normal lung, we show that reintroduction of miR-34c-3p reduces NSCLC cell survival in vitro. Further, in order to deliver the miR-34c-based therapeutic selectively to tumor cells, we took advantage of a reported nucleic acid aptamer (GL21.T) that binds and inhibits the AXL transmembrane receptor and is rapidly internalized in the target cells. By applying methods successfully used in our laboratory, we conjugated miR-34c to the GL21.T aptamer as targeting moiety for the selective delivery to AXL-expressing NSCLC cells. We demonstrate that miR-34c-3p and the GL21.T/miR-34c chimera affect NSCLC cell proliferation and are able to overcome acquired RTK-inhibitor resistance by targeting AXL receptor. Thus, the GL21.T/miR-34c chimera exerts dual inhibition of AXL at functional and transcriptional levels and represents a novel therapeutic tool for the treatment of NSCLC.
Keywords: miRNA, aptamer, NSCLC, lung cancer, therapeutics
Introduction
Despite years of research and progresses in medical treatments, lung cancer remains the leading cause of cancer-related death worldwide.1 According to histology and molecular biology, lung cancer is divided into two main groups: small-cell lung cancer (SCLC) and the more frequent non-small-cell lung cancer (NSCLC), with an expected 5-year survival rate of about 15%.2 This is mainly due to the presence of metastatic disease at the time of diagnosis in many patients. Thus, improvements in survival require more effective therapies. Molecularly targeted therapies in NSCLC patients were initially used in the late 1990s with the introduction of a tyrosine kinase inhibitor (TKI), gefitinib, targeting the epidermal growth factor receptor (EGFR) in NSCLC harboring EGFR activating mutations.3
However, many patients develop a resistance to target therapy, due to newly acquired mutations or to the overexpression of different tyrosine kinase receptors such as AXL.4 It is therefore an urgent need to develop new therapeutic approaches to further improve survival of these patients.
MicroRNAs (miRNAs) are small non-protein-coding RNAs, 19–25 nucleotides in length that negatively regulate gene expression by translational repression or inducing degradation of mRNA targets.5 miRNAs act in concert regulating the intracellular levels of key proteins and have been shown to be deregulated in different human cancers, suggesting their crucial role in cancer pathogenesis.6, 7 For their capability to target many genes, they are emerging as promising RNA-based therapeutics in different diseases, including cancer.8, 9, 10, 11
The tumor-suppressive miR-34c and its cognate miRNAs, miR-34a and miR-34b, have been shown to be regulated by and to mediate p53 suppression of cell cycle entry in S phase, promoting damaged cells to undergo apoptosis.12 Indeed, miR-34c and family members have been consistently found downregulated in several tumors, including NSCLC, allowing cell survival and resistance to apoptosis.13, 14, 15, 16, 17 Despite the growing evidence of the potential of miR-34 as an effective therapeutic for several kinds of cancer, the lack of a selective delivery carrier remains today the key obstacle that limits its translation to the clinic.18 In this regard, nucleic acid-based aptamers represent a very promising tool as carriers for delivering RNA therapeutic molecules to target cells and tissues.19 Aptamers are short single-stranded oligonucleotides (DNA or RNA), that, via assuming specific and complex three-dimensional folding,20 bind with high affinity and specificity to their target molecules.21, 22 Aptamer ligands for transmembrane receptors have been recently generated that are rapidly recycled, with the receptor serving as selective targeting moieties.19, 23
Currently, an increasing number of aptamers targeting cancer cell surface epitopes have been successfully used for the specific delivery of active drug substances both in vitro and in vivo, including nanoparticles,24 anti-cancer therapeutics,25 toxins,26 enzymes,27 radionuclides,28 viruses,29 small interfering RNAs (siRNAs),30 and more recently miRNAs.31
Here, we validated the therapeutic potential of miR-34c-3p for NSCLC and addressed its specific targeted delivery by adopting a non-covalent approach to conjugate a miR-34c mimic to a nucleic acid aptamer, generating an aptamer-miRNA chimera. To this end, we took advantage of a recently characterized 2′-fluoro-pyrimidine-containing RNA aptamer named GL21.T, that upon binding inhibits the AXL receptor and is rapidly internalized into the target cell in a receptor-mediated manner.32 As previously reported, GL21.T has revealed to be an effective and safe mediator for the functional delivery of miRNAs into lung cancer cells, promoting the inhibition of target gene expression.33, 34, 35 Here, we adopted GL21.T for the selective delivery of miR-34c to AXL-expressing NSCLC patient-derived cells, demonstrating a reduction of cell proliferation, migration, and sensitivity to ionizing radiation. Moreover, the GL21.T aptamer-miR-34c chimera induced a recovery of erlotinib resistance in resistant NSCLC cell lines.
Results
miR-34c Is Downregulated in NSCLC Tissues and Cell Lines
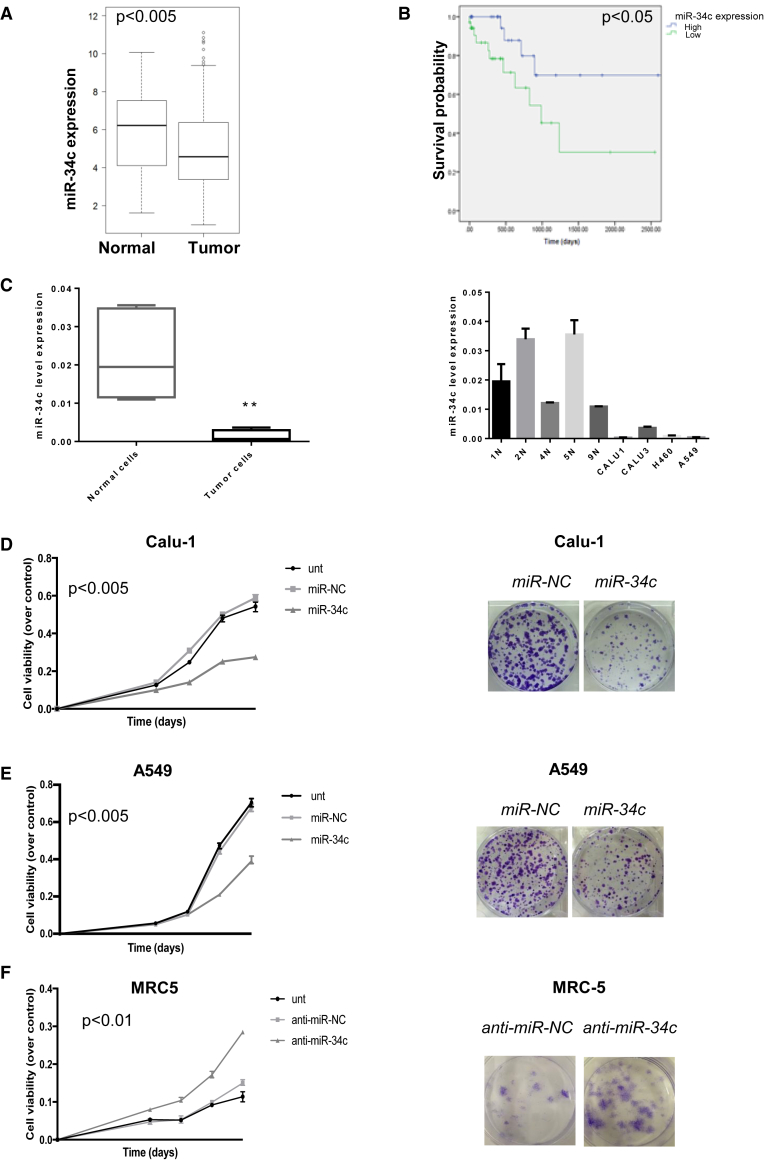
We analyzed miR-34c-5p and -3p expression in a large cohort of NSCLC patients within the Cancer Genome Atlas (TCGA) database (515 NSCLC and 46 normal lung samples) by bioinformatics analysis. As shown, miR-34c levels resulted in significantly downregulated in NSCLC compared to normal lung tissues, with a significantly lower median value (Figure 1A). The log rank Mantel-Cox test showed that patients with higher levels of miR-34c had a longer overall survival (OS), suggestive of a prognostic role for miR-34c (p < 0.05) (Figure 1B).
Figure 1.
miR-34c in Human NSCLC Tissues
(A) Significant increase of miR-34c expression was identified in normal lung versus adenocarcinoma tissues collected from the TCGA database (t test, p < 0,005). (B) Kaplan-Meier survival analysis for TCGA NSCLC patients with high and low miR-34c expression. The survival data were compared using the log rank test (p < 0.05). (C) Expression of miR-34c-3p in several tumor cell lines (Calu-1, A549, Calu-3, and H460) was lower compared to normal primary lung cell lines (1N, 2N, 4N, 5N, and 9N). MiR-34c-3p expression was assessed by real-time PCR. The transcript level was normalized over RNU6B expression, used as an internal reference. Bar graphs indicate mean value ± SD and the p value is calculated by using Student’s t test, **p < 0.01. (D) Calu-1 and (E) A549 cells were transfected with control miR (miR-NC) or miR-34c-3p, and cell proliferation was analyzed by MTS assay 3, 4, 5, and 6 days after transfection (left). Bar graphs indicate mean value ± SD and the p value is calculated by using Student’s t test, p < 0.005 compared to the non-transfected cells (Unt); Calu-1 and A549 cells were transfected with miR-NC or miR-34c-3p. Overexpression of miR-34c-3p significantly inhibited colony formation (right). (F) MTS assay determined cell proliferation in MRC-5 cells following downregulation of miR-34c-3p (left). Bar graphs indicate mean value ± SD and the p value is calculated by using Student’s t test, p < 0.01, compared to non-transfected cells (Unt); colony-formation assay determined the effect of downregulated miR-34c-3p on colony-forming ability in MRC-5 cell lines (right). Downregulation of miR-34c-3p in normal lung cells promotes cell proliferation and allows colony formation.
Next, we analyzed miR-34c-3p expression levels in NSCLC cells (A549, Calu-1, H460, and Calu-3), and in normal cells (patient-derived normal lung cell lines, 1N, 2N, 4N, 5N, and 9N) by qRT-PCR. As shown in Figure 1C, intracellular levels of miR-34c in NSCLC cell lines were significantly lower compared to normal lung cells. These results indicated that miR-34c is consistently downregulated in NSCLC tumors and tumor-derived cell lines, supporting a potential role of this miRNA in NSCLC as a favorable prognostic marker.
Overexpression of miR-34c Inhibits In Vitro Growth of NSCLC Cells
Together with the cognate miRNAs, miR-34a, and miR-34b, miR-34c has been reported to play a tumor-suppressive role in cancer. In order to define the potential role of miR-34c-3p on NSCLC cell proliferation, we took advantage of two low-expressing miR-34c cell lines, Calu-1 and A549. Cells were transiently transfected with either miR-34c-3p or control miR (miR-NC) and analyzed by MTS and colony formation assay. As shown in Figures 1D and 1E (left), increased amounts of miR-34c (Figure S1A) led to reduced cell viability in both cell lines as compared to negative controls (untreated or transfected with miR-NC cells).
We then evaluated the long-term effects of miR-34c-3p on proliferation, performing a colony-formation assay. The colony number of Calu-1 and A549 cells transfected with miR-NC was significantly higher compared to the cells transfected with miR-34c mimic (Figures 1D and 1E, right). To further confirm these data, we evaluated the effects of miR-34c-3p silencing in normal lung MRC-5 cells. As shown, decreased miR-34c expression resulted in a significant increase of cell proliferation and colony formation capability compared to control cells (untreated or transfected with anti-miR-NC) (Figure 1F). All together, these data demonstrated that miR-34c can effectively modulate cell growth.
AXL as a Direct Target of miR-34c
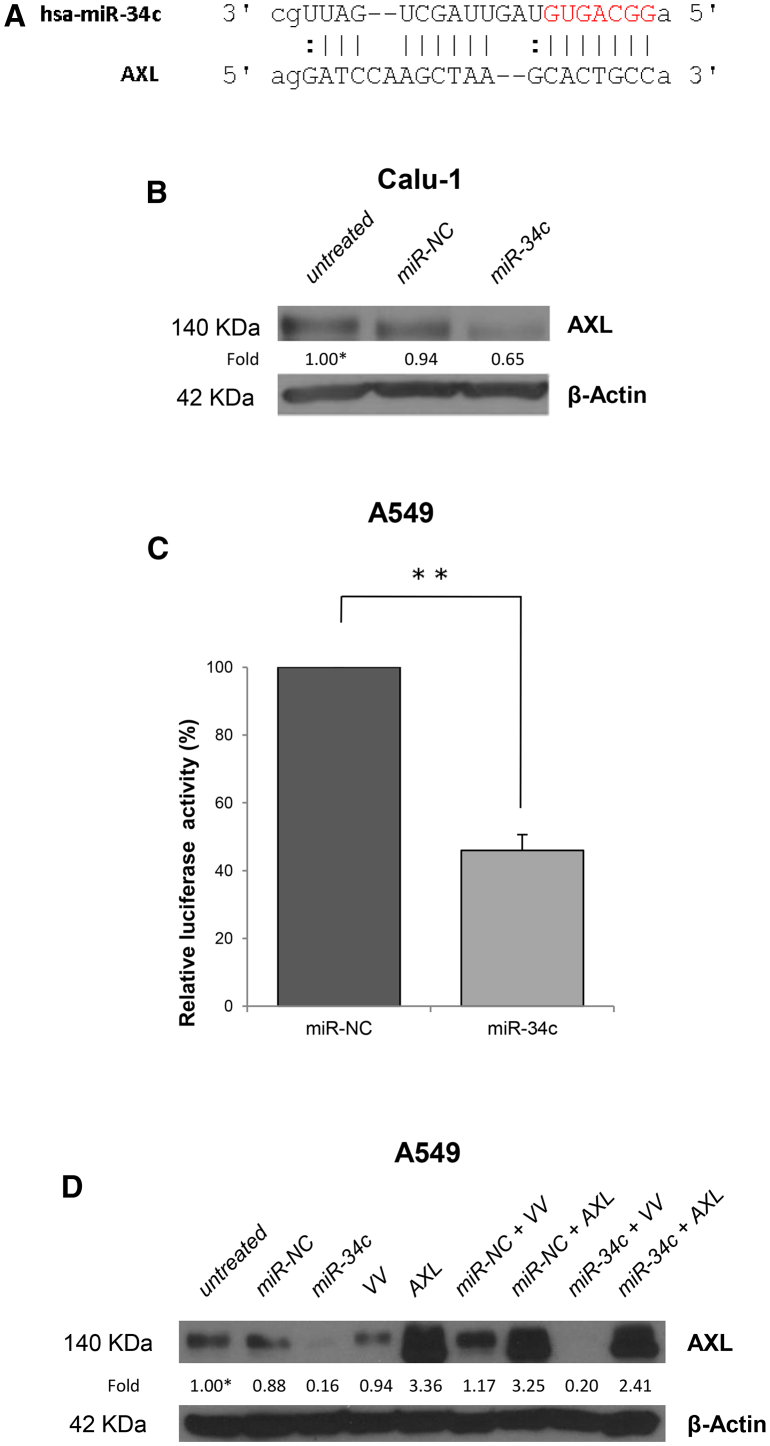
The transmembrane receptor tyrosine kinase, AXL, is a target of miR-34a36, 37 that has been recently shown to play a key role in acquired resistance to EGFR inhibitors in NSCLC.4 We thus verified whether it could be a target also of miR-34c-3p. By using miRNA target prediction algorithms (RNA hybrid), we identified a putative miR-34c-3p binding site located within the 3′ UTR of AXL (Figure 2A). In order to validate the AXL transcript as a target of miR-34c, we determined whether the binding of miR-34c-3p to its 3′ UTR would result in the inhibition of AXL gene expression. To this end, we first examined AXL protein levels in Calu-1 cells upon 72 hr of transfection with pre-miR-34c-3p. As shown in Figure 2B, exogenous miR-34c-3p induced a clear reduction of AXL protein levels by approximately 35% as compared to controls. In addition, in order to validate whether miR-34c directly binds to its predicted site of AXL-3′ UTR mRNA, we conducted a dual luciferase reporter assay for the 3′ UTR of human AXL. To this end, we transiently co-transfected A549 cells with AXL-3′ UTR together with miR-34c-3p. As shown in Figure 2, we observed a significant and consistent reduction in luciferase activity (>50%) at 48 hr of transfection with miR-34c-3p, but not with control miRNA (miR-NC) (Figure 2C).
Figure 2.
miR-34c Targets AXL-3′ UTR and Regulates AXL Expression
(A) The predicted miR-34c-3p binding sites on the 3′ UTR of AXL mRNA (predicted by the RNA HYBRID program). (B) AXL expression was analyzed in Calu-1 cells, untreated or transfected with miR-NC or miR-34c-3p for 72 hr, by western blot analysis. β-actin was used as internal control. (C) A549 cells were transiently transfected with AXL-3′ UTR in the presence of miR-34c-3p or miR-NC. Luciferase activity was evaluated 48 hr after transfection. Bar graphs indicate mean value ± SD and the p value is calculated by using Student’s t test, **p < 0.01. (D) Western blot analysis of AXL protein expression in A549 cells co-transfected with vector control (VV) or AXL plasmid lacking the 3′ UTR region (AXL) and miR-34c-3p or miR-NC. β-actin was used as internal control.
The functional relationship between miR-34c-3p and AXL was confirmed using a rescue strategy after transfection of A549 cells with miR-34c and AXL cDNA plasmid lacking the 3′ UTR region. AXL protein levels were detected by western blot. Collectively, AXL and miR-34c-3p, but not the 3′ UTR deletion mutant, rescued AXL protein levels (Figure 2D), suggesting that miR-34c-3p may regulate, at least in part, cell growth of NSCLC cells by targeting AXL.
Design and Folding of an Aptamer-miRNA Conjugate
The development of miRNA selective delivery strategy is a key aspect for their therapeutic application. To address this issue, we generated, via stick-end annealing, a molecular aptamer-miRNA chimera (termed GL21.T-miR-34c) consisting of a duplex miRNA cargo and a nucleic acid aptamer as delivery carrier. We fused the miR-34c to the GL21.T aptamer that selectively binds and inhibits the AXL receptor using complementary stick sequences linking the GL21.T aptamer and the miRNA passenger strand. Finally, we annealed the guide strand of miRNA to the template (Figure S1B). Based on our previous report,34 we used as miRNA mimetic the distal stem portion of the human miRNA-34c encompassing 24 bases of the 5′ strand and 22 of the 3′ strand, derived from the miR-34c precursor, rather than the sequence of the mature miRNA. This would allow the introduction of internal partial complementarity and the production of a more effective Dicer substrate.37
We first verified the correct annealing of the conjugate by the presence of a shifted band of migration by nondenaturing gel electrophoresis analysis (Figure S1C, lane 4).
Then, we analyzed GL21.T-miR-34c binding ability on A549 (AXL+) and, as shown in Figure S1D, the conjugate preserves the binding capability compared to the GL21.T alone.
To test whether the aptamer-conjugated miRNA was recognized and correctly processed by Dicer, we incubated GL21.T-miR-34c conjugates in the presence or absence of recombinant human Dicer and analyzed them with non-denaturing gel electrophoresis. As anticipated, the conjugates were recognized and digested by Dicer, producing two cleaved products of the expected molecular sizes, corresponding to the cleaved miRNA and the aptamer alone (Figure S1E).
GL21.T-Mediated Selective Delivery of Functional miR-34c
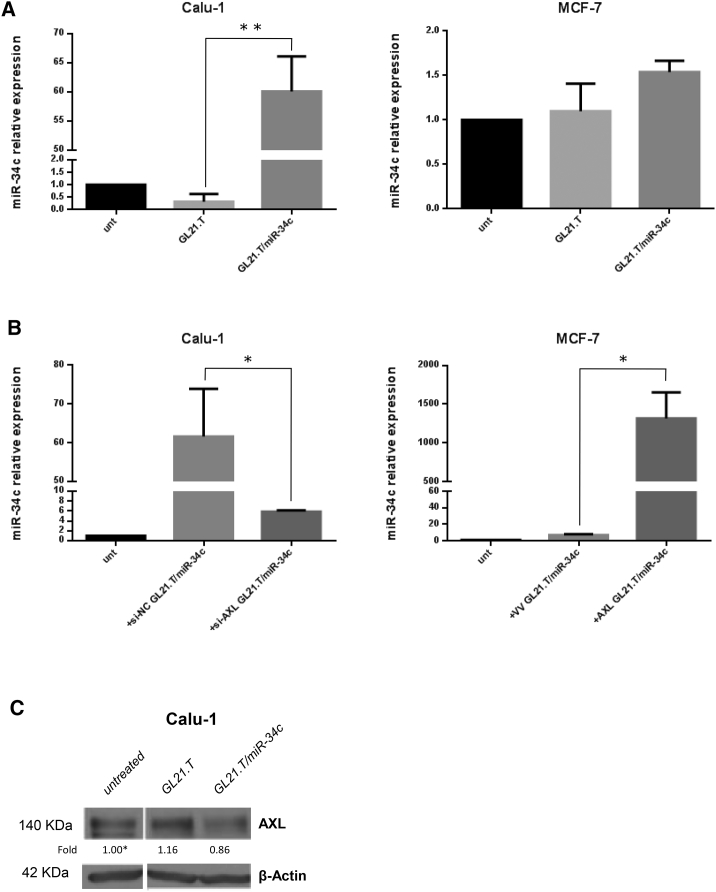
To investigate whether GL21.T aptamer could act as a selective carrier for the delivery of the conjugated miRNA, we performed in vitro experiments with two different cell lines, Calu-1 and MCF-7, expressing high and low levels of AXL protein, respectively (Figure S2A). Calu-1 and MCF-7 cells were treated with GL21.T/miR-34c for 72 hr. Treatment was performed at a concentration of 400 nM since we have previously demonstrated that this concentration assures an efficient and functional GL21.T-mediated miR delivery.34 As expected, qRT-PCR showed that the GL21.T/miR-34c conjugate increased intracellular miR-34c levels in Calu-1 cells (AXL+) (Figure 3A, left panel) but not in MCF-7 cells (AXL) (Figure 3A, right panel).
Figure 3.
Synergistic Effects of the GL21.T-miR-34c Chimera
(A) Calu-1 and MCF-7 cells were treated with GL21.T or GL21.T/miR-34c (400 nM). After 72 hr, miR-34c was quantified by RT-PCR. Bar graphs indicate mean value ± SD and the p value is calculated by using Student’s t test, **p < 0.01. (B) Calu-1 cells were transfected with si-AXL or siRNA control (si-NC) for 48 hr; MCF-7 cells were transfected with AXL or control vector (VV) for 24 hr and treated with GL21.T/miR-34c for 72 hr. miR-34c levels were quantified by RT-PCR. Bar graphs indicate mean value ± SD and the p value is calculated by using Student’s t test, *p < 0.05. (C) Calu-1 cells were treated as indicated, and AXL protein expression was analyzed by western blot after 72 hr. β-actin was used as internal control.
To demonstrate receptor-dependent delivery of miR-34c, we silenced AXL in Calu-1 (AXL+) cells with a specific si-AXL-RNA (Figure S2B, right). Cell transfected for 48 hr with si-AXL-RNA, were incubated with GL21/miR-34c (72 hr), and miR-34c levels transcripts were evaluated by qRT-PCR. As expected, no significant upregulation of miR-34c was detected in Calu-1 cells following GL21.T/miR-34c in the presence of si-AXL as compared to the control (Figure 3B, left). Conversely, the forced expression of AXL in MCF-7 cells (Figure S2B, left) induced an increase of miR-34c level upon chimera treatment (Figure 3B, right). These results indicate that GL21.T can deliver miR-34c inside the cells in an AXL-dependent manner.
As shown above, miR-34c targets AXL-3′ UTR. Accordingly, GL21.T/miR-34c treatment, but not the aptamer alone, suppressed the expression of AXL protein (Figure 3C), establishing a negative feedback loop on AXL expression.
GL21.T/miR-34c Effect on Cell Growth and Migration
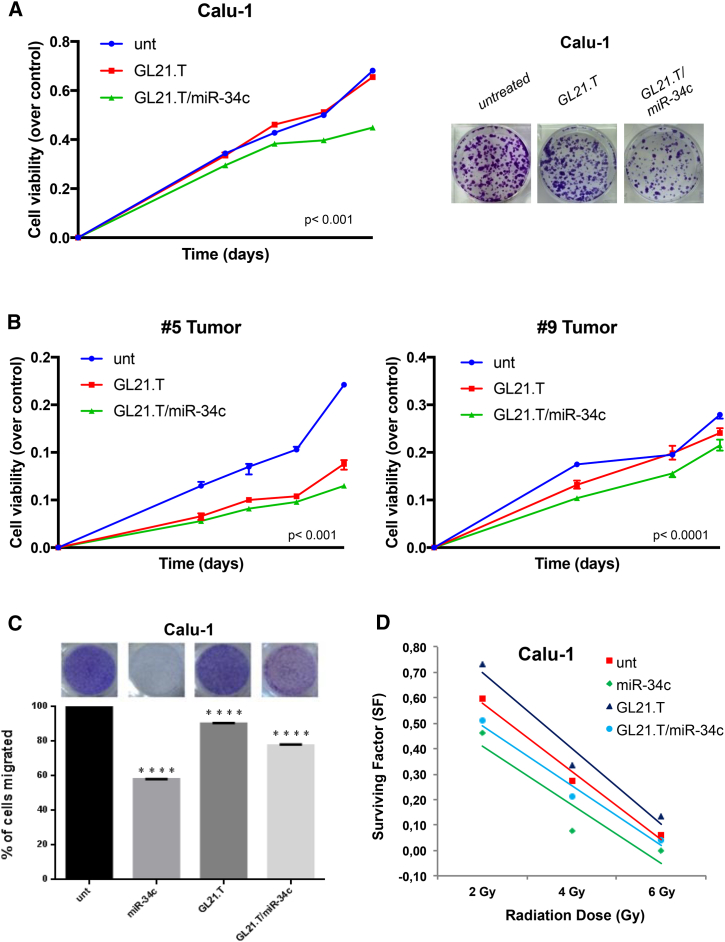
To further support the efficacy of the treatment, we investigated whether miR-34c and the AXL aptamer could cooperate to produce synergistic effects and control cell growth. To this aim, we treated Calu-1 cells with GL21.T/miR-34c conjugate and then analyzed the effects on proliferation. MTS assay showed that cell proliferation of Calu-1 cells was inhibited by GL21.T/miR-34c treatment to almost the same extent as observed after miR-34c transfection (Figure 4A, left panel). Furthermore, the ability to form colonies was impaired when Calu-1 cells were treated with GL21.T/miR-34c (Figure 4A, right panel).
Figure 4.
Role of GL21.T/miR-34c on NSCLC Cell Proliferation
(A) Calu-1 cells were treated with GL21.T/miR-34c and GL21.T aptamer alone. Cell proliferation was analyzed by MTS assay 3, 4, 5, and 6 days upon transfection (left). p < 0.0001, compared to the non-transfected cells (Unt). Cells were treated with GL21.T/miR-34c and GL21.T aptamer alone and colony-formation assay was performed (right). (B) #5T (AXL+) or #9T (AXL−) cells were treated with GL21.T/miR-34c or GL21.T aptamer alone. Cell viability was analyzed by MTS assay. Bar graphs indicate mean value ± SD and the p value is calculated by using Student’s t test, p < 0.001 and < 0.0001, respectively, compared to the non-transfected cells (Unt). (C) Calu-1 cells were transfected with miR-34c-3p or treated with GL21.T aptamer or GL21.T/miR-34c conjugate. The cell migration capability was analyzed by transwell migration assay using 10% FBS as migration inducer. Migrated cells were stained with crystal violet and photographed. ****p < 0.0001, compared to the non-transfected cells (Unt). (D) Effect of GL21.T/miR-34c conjugate on long-term survival of Calu-1 cells after irradiation at different doses.
Since cell lines may not fully recapitulate human cancer, we determined the effects of GL21.T/miR-34c aptamer-miRNA chimera in primary cells obtained from surgical specimens from lung cancer patients. We first analyzed AXL protein levels in different primary lung cancer cells to discriminate patients with high and low levels of AXL expression (Figure S2C). Next, we analyzed the effect of the aptamer-miRNA chimera on the proliferation of two primary lung cancer models: AXL+ (#5T) and AXL− (#9T). Notably, only AXL+ cells displayed reduced cell proliferation when challenged with the aptamer-miRNA chimera (Figure 4B).
To extend the functional characterization, we assessed the ability of the conjugate to affect cell migration. As shown in Figure 4C, miR-34c transfection and GL21.T/miR-34c treatment inhibited Calu-1 cell migration by approximately 40% and 20%, respectively, compared to untreated cells. Conversely, the migration of Calu-1 cells treated with aptamer alone was similar to that of untreated cells. Moreover, the results of the transwell migration assay showed that high levels of miR-34c could significantly suppress the migration abilities of Calu-1 cells. Since occurrence of radiation resistance is common in NSCLC patients, it is crucial to identify molecules capable of increasing radiosensitivity. Therefore, we evaluated the long-term irradiation effects on cell survival. In Calu-1 cells, overexpression of miR-34c-3p increased the sensitivity of cells to all ionizing radiation doses when compared to control cells. Moreover, GL21.T/miR-34c was able to increase sensitivity to low exposure doses (Figure 4D).
Role of miR34c on Erlotinib Resistance
Recent reports suggest that in NSCLC, AXL overexpression and activation is associated with causal events for the acquired resistance to the epidermal growth factor inhibitor (EGFR-TKI), erlotinib.4 Therefore, we assessed the role of miR-34c-3p and of the chimeric molecule on erlotinib sensitivity in parental HCC827 cells or erlotinib-resistant (ER3) HCC827 cells. AXL levels were confirmed by western blot and by qRT-PCR (Figure S3A). As expected, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay revealed that ER3 cells did not show any significant reduction of cell viability upon erlotinib incubation at different times and doses (Figure S3B). AXL knockdown was able to partially restore erlotinib sensitivity (Figures S3C and S3D), suggesting that drug resistance is in part due to AXL-mediated signaling. To assess the putative role of miR-34c-3p in erlotinib sensitivity, ER3 cells were transfected with miR-34c-3p (or miR-NC), and cell proliferation was analyzed. As shown in Figure 5A, the number of viable cells was reduced upon miR-34c transfection compared to control (untreated cells or transfected with miR-NC), and these changes were further increased after the erlotinib treatment.
Figure 5.
The Role of miR-34c on Erlotinib Resistance
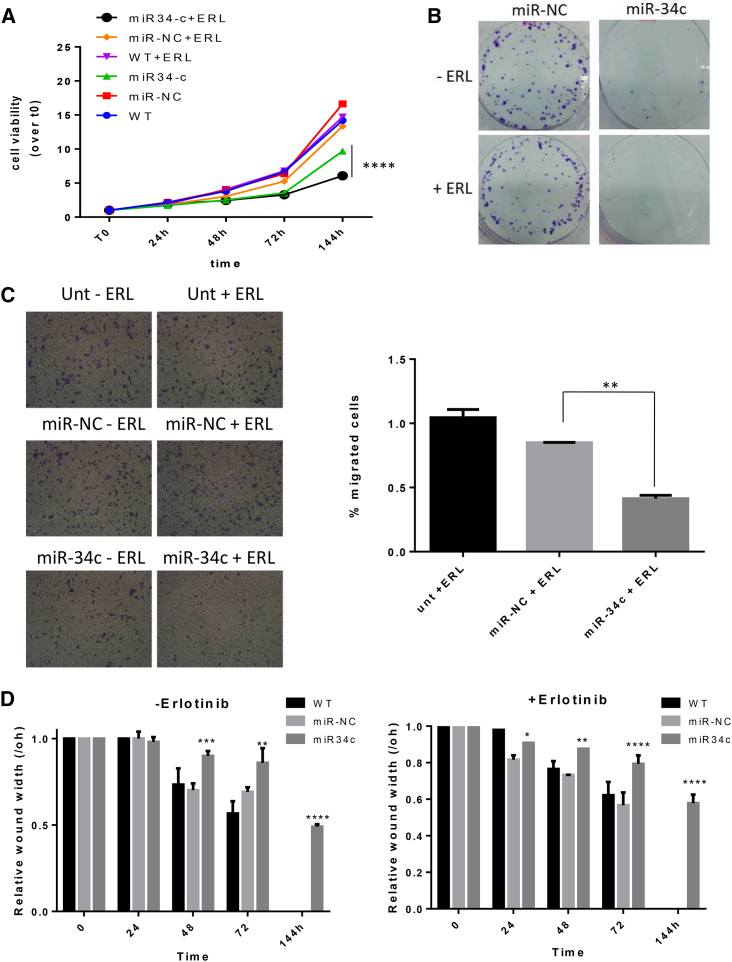
(A) ER3 cells were transfected with miR-34c-3p or miR-NC. After 72 hr, transfected ER3 cells were seeded in 96-wells and treated or not with erlotinib. Cell proliferation was measured by the MTT assay 1, 2, 3, and 6 days after transfection. (B) ER3 cells were transfected with miR-34c-3p or miR-NC. After 72 hr, transfected ER3 cells were treated or not with erlotinib. Overexpression of miR-34c-3p restores sensitivity to the drug by reducing cell proliferation and colony formation. (C) Cell migration assay of ER3 cells upon miR-34c-3p and erlotinib treatment. Cells transfected with scrambled miRNA or miR-34c-3p were harvested after 72 hr and cultured on a membrane (8.0 μm pore size) inserted in the wells of a 24-well plate. Percentage of migrated cells was evaluated by eluting crystal violet solution with 1% SDS and reading the absorbance at 570 nm. Data were obtained from three independent experiments and bar graphs indicate mean value ± SD and the p value is calculated by using Student’s t test, *p < 0.05; **p < 0.01; ***p < 0.001 (over control). (D) Wound-healing assays. ER3 cells were transfected with miR-34c-3p for 72 hr and then seeded into 6-well plates at 80%–90% confluence and treated for 72 hr with GL21.T/miR-34c molecule. Bar graphs indicate mean value ± SD and the p value is calculated by using two-way ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 (over control).
The long-term effect on proliferation was then assessed with a colony assay. ER3 cells transfected with miR-34c-3p and treated with erlotinib exhibited lower numbers of colonies compared to negative control cells (untreated cell and cell transfected with miR-NC) (Figure 5B).
miR-34c Affects Cell Migration of ER3 Cells
As tumor invasion and metastatic potential are linked to cell migration, we then evaluated whether miR34c could affect erlotinib’s effects on cell migration. We found that miR-34c-3p inhibited ER3 migration when compared to cells transfected with miR-NC (Figure 5C). Moreover, when ER3 cells were treated with erlotinib, migration was further reduced (∼60%). These findings were confirmed with multiple scratch assays (Figures 5D and S4).
GL21.T-mir34c Affects Erlotinib Resistance
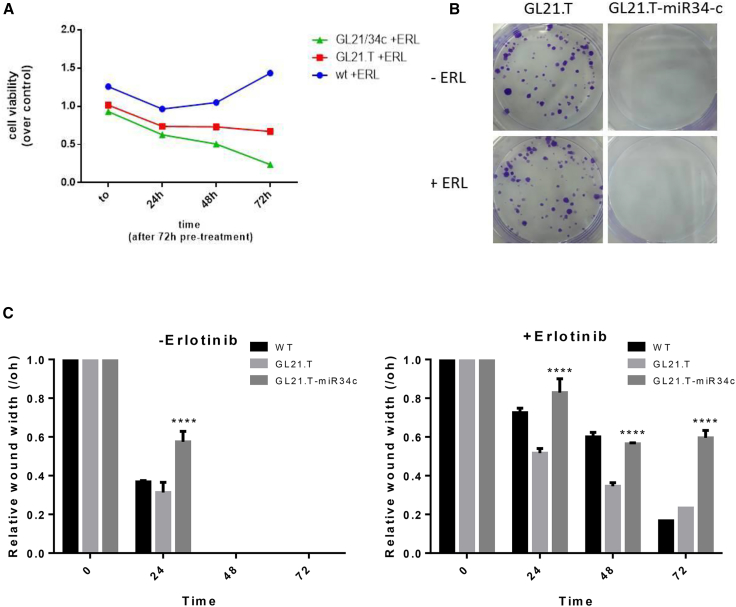
Given the role of miR-34c-3p on erlotinib resistance, we tested GL21.T/miR34c ability to preserve this function. To this end, AXL-expressing ER3 cells were treated with GL21.T or with GL21.T/miR34c chimera in the presence of erlotinib, and the effects on cell proliferation determined. Compared to untreated cells, the number of viable cells was reduced upon treatment with the anti-AXL GL21.T aptamer and further reduced upon aptamer-miRNA chimera treatment (Figure 6A). Next, we evaluated long-term effects of aptamer-miRNA chimera treatment on proliferation of erlotinib-resistant ER3 cells. As assessed by colony-formation assay, erlotinib treatment could significantly reduce the number of ER3 colonies in the presence of GL21.T/miR-34c aptamer-miRNA chimera (Figure 6B). Scratch assay confirmed these findings (Figures 6C and S5).
Figure 6.
GL21.T/miR-34c Affects Erlotinib Resistance
(A) ER3 cells were treated with 400 nM of GL21.T aptamer or GL21.T/miR34c chimera for 72 hr and treated with erlotinib. Cell proliferation was measured by the MTT assay. (B) ER3 cells were treated for 72 hr with 400 nM GL21.T or GL21.T/miR34c, renewing treatment after 72 hr and treated or not with erlotinib. Long-term proliferation was determined by colony-formation assay. (C) A wound of approximately 1 mm in width was scratched with a 20-μL pipette tip. Wound closure was monitored at the indicated time intervals and imaged with phase contrast microscopy on an inverted microscope (Olympus 1 × 51 using a 5 × phase contrast objective). Bar graphs indicate mean value ± SD, and p is calculated by using two-way ANOVA. ****p < 0.0001 (over control).
Serum Stability of GL21.T-34c
An important feature for clinical translation of new therapeutics is their stability in the circulation. Therefore, to evaluate the resistance of aptamer-miRNA chimera to degradation, we incubated the conjugate with human serum at different intervals, up to 96 hr. Serum-RNA samples were recovered (0, 1, 2, 4, 8, 24, 48, 72, and 96 hr) and analyzed by non-denaturing polyacrylamide gel electrophoresis (Figure S1F), demonstrating that GL21.T-miR-34c conjugate is stable up to approximately 8 hr and then is gradually degraded.
Discussion
Treatment of NSCLC-expressing mutated EGFR with EGFR inhibitors, although effective for most patients, is hampered by occurrence resistant clones due to de novo or increased expression of alternative RTKs, including AXL.38 Therefore, the development of RNA therapeutics that interfere with RTK expression, restoring drug sensitivity specifically to target cancer cells, would increase the effectiveness of NSCLC treatments. Yet, the biggest challenge in siRNA therapy (siRNA and miRNA) remains efficient and specific delivery to the desired target tissue. In this regard, nucleic acid-based aptamers have been shown to be very promising tools for delivery of therapeutic cargos.19
In this study, we deepen the role of miR-34c in NSCLC and an aptamer-miRNA conjugate (GL21.T/miR-34c), non-covalently joining GL21.T, an anti-AXL aptamer, to the tumor suppressor miR-34c-3p. We demonstrate that the aptamer-miRNA chimera is able to selectively target AXL-expressing cells, rescuing sensitivity of NSCLC to erlotinib and leading to cell death.
Among all downregulated miRNAs reported for NSCLC,39, 40, 41, 42, 43, 44 we focused our attention on miR-34c, a tumor suppressor miRNA largely investigated in different types of cancers.17, 45, 46, 47
We first evaluated miR-34c expression in NSCLC tissues compared to non-tumor lung tissues, in data collected from TCGA database. MiR-34c resulted in significantly less expressed in NSCLC samples compared to normal lung. More importantly, in a cohort of NSCLC patients (64 primary specimens from TCGA), statistical analysis revealed that patients with higher expression of miR-34c showed significantly longer survival and better prognosis. Taken together, these data suggest that miR-34c downregulation might be linked to poor outcome. Further, we provided evidence for the first time in NSCLC cells that AXL transcript is a direct target and its translation is regulated by miR-34c-3p. This finding is of importance in light of recent reports showing that elevated AXL levels induce acquired resistance to treatment with EGFR inhibitors.4 Accordingly, we demonstrated the ability of miR-34c-3p rescue sensitivity to erlotinib of NSCLC-derived cells (ER3) that were selected for acquired resistance to erlotinib. We showed that, likely by downregulating AXL, the miR-34c-3p is able to restore sensitivity to erlotinib treatment, reducing ER3 cell proliferation and migration.
Despite the therapeutic potential of miR-based molecules, the development of efficient delivery strategies is a key aspect for their development.
We therefore designed an anti-AXL aptamer-miRNA chimera, combining the targeting and antagonist functions of GL21.T with the silencing potential of miR-34c.
As previously reported for other aptamer-miRNA chimeras with anti-RTK aptamers, we show that GL21.T serves as a selective targeting moiety for the intracellular uptake of miR-34c.48 Following treatment with GL21.T/miR-34c, we observed miRNA uptake into AXL-expressing target tumor cells (Calu-1), but not in cells that do not express AXL (MCF-7). Aptamer-mediated miRNA overexpression resulted in AXL downregulation and in decreased cell proliferation, as also observed in cells transfected with the miRNA. Similar results on cell proliferation were also observed in experiments performed in primary lung cancer cells obtained from human lung biopsies. These results correlate with previous reports demonstrating miR-34c-3p to function as tumor suppressor by inhibiting multiple pathways involved in cancer growth. Zhou et al.13 demonstrated that overexpression of miR-34c-3p suppressed cell proliferation and migration of A549 cells by targeting PAC1. Liu et al.49 demonstrated that miR-34c-3p expression was significantly reduced in tissues and serum samples from NSCLC patients and in NSCLC cell lines and was able to modulate cell proliferation and migration by targeting eIF4E.
We also demonstrated that the conjugate acts in a cell-specific manner. Indeed, we show that both the amounts of intracellular miR-34c and the extent of target downregulation were dependent upon the level of expression of AXL on the cell surface.
Most importantly, we showed that the aptamer-miRNA chimera preserves as well the ability to restore sensitivity to erlotinib treatment. Our results thus suggest the therapeutic potential of a novel combined strategy in which the use of the aptamer-miRNA chimera by downregulating miR-34c target gene including AXL enhances the cell responsiveness of erlotinib. Consequently, although the downregulation of AXL would reduce cell responsiveness to the aptamer-miRNA chimera treatment, the combined protocol by restoring cell sensitivity to erlotinib can induce cell death, ultimately leading to a complete inhibition of tumor growth.
In conclusion, the GL21.T/miR-34c aptamer-miRNA conjugate is the first example of selective targeted delivery of miRNAs able to overcome acquired RTK-inhibitor resistance in NSCLC. In addition to NSCLC, the occurrence of acquired resistance to EGFR inhibitors and tumor relapse is frequent in other solid tumors, such as breast. Therefore, the designed aptamer-miRNA chimera has a broader applicability to several cancer types where it would be beneficial to increase drug responsiveness while reducing toxicity associated with systemic treatments.
Materials and Methods
TCGA miRNA Dataset and Patient Information
miR34c-5p and -3p expression data and corresponding clinical information for NSCLC datasets were downloaded from the TCGA data portal in June 2013 (https://tcga-data.nci.nih.gov/). The collection of the data from the TCGA platform was compliant with laws and regulations for the protection of human subjects, and necessary ethical approvals were obtained.
The level 3 data for the 218 NSCLC patients were quantile normalized and log2 transformed. Only patients with OS information were taken into account.
The association between continuous miRNA expression and OS was carried out using univariable Cox regression analysis. The difference in the survival outcome between risk groups was estimated by log rank Mantel-Cox test and plotted by Kaplan-Meier curve. The analyses were performed with BRB-Array Tools-R/BioConductor (version 2.10). For the multivariable analysis, the Cox proportional hazard model was applied, and a backward stepwise selection procedure (Wald) was used to identify miRNAs with independent prognostic value. All reported p values were two-sided. The analyses were performed using SPSS (version 2.1).
Cell Lines
MRC-5, Calu-1, Calu-3, and MCF-7 cells were cultured under standard conditions in DMEM, while A549, H460, ER3, and HCC827 cells were grown in RPMI-1640 medium, all supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin. All media and supplements were from Sigma-Aldrich (Milan, MI, Italy). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. All NSCLC cell lines were purchased from American Type Culture Collection (ATCC). A549, MCF-7, and HCC827-ER3 cells were kindly provided by Dr. Balazs Halmos (Columbia University Medical Center, New York, NY, USA).
Isolation of Primary Cell Cultures from Human Lung Biopsies
Human lung biopsies (samples from Azienda Ospedaliera Universitaria Primo Policlinico, Naples, NA, Italy) were cut by mechanical fragmentation with sterile scissors and tongs. Isolated cells were routinely seeded onto 100-mm dishes and grown in DMEM-nutrient F12-Ham (DMEM-F12). Media were supplemented with 10% heat-inactivated FBS and 100 U/mL penicillin and streptomycin. All media and supplements were from Sigma-Aldrich (Milan, MI, Italy). Primary cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Ethical permission was obtained from the Ethics Committee of Federico II University, Naples, Italy.
Cell Transfection
For transient transfection with miRNA-34c cells were seeded in 6-well plates 1 day ahead, grown to 50%–70% confluence and then transfected with 100 nM (final concentration) of pre-miR-34c-3p (cat. #MS00009548, Ambion, Life Technologies, Monza, MB, Italy), pre-miR-negative control #1, anti-miR-34c or anti-control miR (Ambion, Life Technologies, Monza, MB, Italy, cat. #AM17000), using Oligofectamine (Invitrogen, Life Technologies, Monza, MB, Italy), according to the manufacturer’s protocol.
To transiently knock down AXL gene expression, si-AXL or siRNA control (Santa Cruz Biotechnologies, D.B.A. Italia, Segrate, Milan, MI, Italy) were transfected using oligofectamine at a final concentration of 50 nM (Santa Cruz Biotechnologies, D.B.A. Italia, Segrate, Milan, MI Italy). For transient overexpression of AXL cDNA TruClone (4 μg, final concentration) (Origene, Rockville, MD, USA), or 3′ UTR AXL plasmid (InvivoGen), cells were transfected using Lipofectamine 2000 (Invitrogen, Life Technologies, Monza, MB, Italy), according to the manufacturer’s protocol.
Aptamer-miRNA Chimera
The following sequences were used for chimera production: GL21.T sticky, 5′-AUGAUCAAUCGCCUCAAUUCGACAGGAGGCUCACXXXXGUACAUUCUAGAUAGCC-3′; miR-34c guide, 5′-AAUCACUAACCACACGGCCAGG-3′; and miR-34c passenger sticky, 5′-ACUAGGCAGUGUAGUUAGCUGAUUGC/GGCUAUCUAGAAUGUAC-3′.
All RNAs were modified with 2′-F pyrimidines and synthesized by TriLink Biotechnologies (San Diego, CA, USA). Stick sequences, consisting of 2′-F-Py and 2′-OMe purines, are underlined. The italic X indicates a covalent spacer, a three-carbon linker ((CH2)3).
Before each treatment, aptamer was subjected to a short denaturation-renaturation step (5 min 85°C, 2 min on ice, 10 min at 37°C). To prepare GL21.T/miR34c, 5 μM of miR-34c strands (miR-34c guide and miR-34c passenger sticky) were incubated at 95°C for 15 min, 55°C for 10 min, and room temperature (RT) for 20 min, in Binding Buffer 10× (200 mM HEPES [pH 7.4], 1.5 M NaCl, 20 mM CaCl2). 5 μM of sticky aptamer was then annealed by incubation at 37°C for 30 min.
For all the experiments, treatments with GL21.T aptamer or GL21.T/miR34c chimera were performed at 400 nM (final concentration).
Cell Binding by qRT-PCR
Cells were seeded in 3.5-cm plates and treated with 200 nmol/L of GL21.T-34c or GL21.T, used as bond control, for 30 min at 37°C. To check the cell binding, following three washes with PBS to remove unbound RNA, the bound RNA was recovered by Trizol (Life Technologies) containing 0.5 pmol/mL of CL4 aptamer (CL4, 5′-GCCUUAGUAACGUGCUUUGAUGUCGAUUCGACAGGAGGC-3′), used as a reference control. The amount of bound RNAs was determined by performing qRT-PCR, as reported,33 with the following primers: GL21 (forward), 5′-AGATCATGATCAATCGCC-3′; GL21 (reverse), 5′-GTGAGCCTCCTGTCGA-3′; CL4 (forward), 5′-GCCTTAGTAACGTGCTTT-3′; and CL4 (reverse) 5′-GCCTCCTGTCGAATCG-3′. The obtained data were normalized to the CL4 reference control. The bond has been expressed as fold change to GL21.T aptamer.
RNA Extraction and Real-Time PCR
Total RNAs (miRNA and mRNA) were extracted using Trizol reagent (Invitrogen, Life Technologies, Monza, MB, Italy), according to protocols recommended by the manufacturer.
Reverse transcription of total miRNA was performed starting from equal amounts of total RNA/sample (500 ng) using miScript reverse transcription kit (QIAGEN, D.B.A. Italia, Segrate, MI, Italy). Quantitative analysis of miR-34c and RNU6B (as internal reference) was performed by real-time PCR using specific primers (QIAGEN, D.B.A. Italia, Segrate, MI, Italy) and miScript SYBR Green PCR Kit (QIAGEN, D.B.A. Italia, Segrate, MI, Italy). The reaction for detection of miRNAs was performed as follows: 95°C for 15’, 40 cycles of 94°C for 15’’, 55°C for 30’’, and 70°C for 30’’.
For reverse transcription of mRNA SuperScript III Reverse Transcriptase (Invitrogen, Life Technologies, Monza, MB, Italy) was used. Quantitative analysis of AXL and β-actin (as internal reference) was performed by real-time PCR using specific primers and iQ SYBR Green Supermix (Bio-Rad, Segrate, MI, Italy). The reaction for detection of mRNAs was performed as follows: 95°C for 15’, 40 cycles of 94°C for 15’’, 57°C for 30’’, and 72°C for 30’’. All reactions for detection of mRNAs or miRNAs were run in a final 25 μL volume.
The threshold cycle (CT) was defined as the fractional cycle number at which the fluorescence passes the fixed threshold. For quantization, the 2(−ΔΔCT) method was used.50 Experiments were carried out in triplicate for each data point, and data analysis was performed with Applied Biosystems StepOne Plus Real-Time PCR Systems.
Protein Isolation and Western Blotting
Cells were washed twice in ice-cold PBS and lysed in lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 5 mM EGTA, 1 mM Na3VO4, and 1× protease inhibitor cocktail).
Protein concentration was determined by the Bradford assay (Bio-Rad, Segrate, MI, Italy) using BSA as the standard, considering that 5 μg of BSA shows a value of absorbance of 0.333 at a wavelength of 595 nm. Equal amounts of protein were denatured in sample buffer (10% SDS, 87% glycerol, 5% β-mercaptoethanol and 0.1% bromophenol blue) for 5 min at 100°C, and then analyzed by SDS-PAGE (10% acrylamide).
Gels were electroblotted onto nitrocellulose membranes (G&E Healthcare, Milan, MI, Italy) and, after the transfer, membranes were blocked for 1 hr with a solution of 5% non-fat dry milk in Tris-buffered saline (TBS) containing 0.1% Tween 20.
Filters were probed with the primary antibodies with shaking overnight at 4°C. Detection was performed by peroxidase-conjugated secondary antibodies using the enhanced chemiluminescence system (Thermo Fisher, Life Technologies, Monza, MB, Italy). Primary antibodies used were anti-AXL (R&D Systems, Milan, MI, Italy) and anti-β-actin (Sigma Aldrich, Milan, MI, Italy).
In Vitro Proliferation Assay
Cell viability was evaluated with the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Milan, MI, Italy), according to the manufacturer’s protocol. The assay is based on reduction of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) to a colored product that is measured spectrophotometrically, as optical density.
Cells were transfected with miRNAs (or anti-miRNAs) or treated with chimera or aptamer alone for 24 hr. Subsequently, cells were trypsinized, plated in 96-well plates in triplicate, and incubated at 37°C in a 5% CO2 incubator.
Metabolically active cells were detected by adding 20 μL of MTS reagent to each well. After 30 min of incubation, the plates were analyzed on a Multilabel Counter (Bio-Rad, Segrate, MI, Italy). The optical density is related to the percentage of viable cells.
Colony-Formation Assay
Colony-formation assay was performed for studying the ability of a single cell to grow into a colony. Cells were first transfected with miRNAs (or anti-miRNAs) or treated with chimera or aptamer alone for 24 hr, and then they were plated in duplicate in 6-well plates. After incubation for 1 week at 37°C in humidified 5% CO2, colonies composed of at least 50 cells were visualized by staining with 0.1% crystal violet in 25% methanol solution for 30 min at 4°C. Dishes were washed with water and then left to dry on a bench.
Luciferase Reporter Assay
A549 cells (120,000 cells per well) were seeded in 6-well plates and co-transfected with 1 μg of 3′ UTR AXL plasmid (and miR-34c or control miR), using Lipofectamine 2000. Both firefly and Renilla luciferase expression was measured 48 hr post-transfection, using the Dual Luciferase Assay (Promega, Milan, MI, Italy), according to the manufacturer’s instructions. Three independent experiments were performed in triplicate. 3′ UTR AXL plasmid, in which the Renilla luciferase gene was fused to the 3′ of the AXL gene, was kindly provided by Konrad E. Huppi (National Cancer Institute, Bethesda, USA).36
Rescue Experiments
To determine whether AXL mediates the effects of miR-34c, rescue experiments were performed overexpressing a deletion mutant of AXL lacking the 3′ UTR.
A549 cells were transfected with miR-34c and with the mutant AXL lacking the 3′ UTR using X-tremeGENE 9 DNA Transfection Reagent (Roche), as described. Protein levels were then analyzed by western blotting.
In Vitro Dicer Assay
GL21.T or GL21.T/miR34c (1 μg) were left undigested or digested using Recombinant Human Turbo Dicer Kit (Genlantis, San Diego, CA, USA) according to the supplier’s instructions. RNAs were then loaded on a 15% non-denaturing polyacrylamide electrophoresis gel, stained with ethidium bromide, and visualized with GEL.DOC XR (Bio-Rad) gel camera.
Chimera Stability in Human Serum
GL21.T-miR34c chimera (4 μM) was incubated in 80% human serum for 1 to 96 hr. Type AB Human Serum provided by Euroclone (Cat. ECS0219D) was used. At each time point, 4 μL (16 pmol RNA) was withdrawn and incubated for 1 hr at 37°C with 3 μL of proteinase K solution (600 mAU/mL) in order to remove serum proteins that interfere with electrophoretic migration. Following proteinase K treatment, 9 μL TBE 1× and 2 μL gel loading buffer (Invitrogen, Waltham, MA, USA) were added to samples that were then stored at −80°C. All time point samples were separated by electrophoresis on 10% non-denaturing polyacrylamide gel. The gel was stained with ethidium bromide and visualized with a GEL.DOC XR (Bio-Rad) gel camera.
Transwell Migration Assay
Transwell Permeable Supports, 6.5-mm diameter inserts, 8.0-μM pore size, polycarbonate membrane (Corning Incorporated, Euroclone Spa, Pero, MI, Italy) were used to perform migration assay.
Cells were transfected or treated for 24 hr, re-suspended in serum-free medium, and plated on the upper chamber of a 24-well transwell (Corning Incorporated, Euroclone Spa, Pero, MI, Italy). The lower chamber of the transwell was filled with 600 μL of culture medium containing 10% FBS.
Cells were incubated at 37°C for 20–24 hr. The transwells were then removed from the 24-well plates and stained with 0.1% crystal violet in 25% methanol. Percentage of migrated cells was evaluated by eluting crystal violet solution with 1% SDS and reading the absorbance at 570-nm wavelength.
Clonogenic Cell Survival Assay
The clonogenic assay was performed on Calu-1 cells (transfected or treated), seeded in 6-well plates and incubated for 12 days after irradiation at different doses (0, 2, 4, and 6 Gy). Following the incubation, media were removed and colonies with at least 50 cells were washed with PBS and stained with 0.1% of crystal violet solution. Plate efficiency (PE) was calculated by dividing the number of colonies formed by the number of cells plated. The survival factor (SF) was calculated by dividing the PE of irradiated samples by the PE of the respective non-irradiated samples. SF values were plotted as a function of radiation dose.
In Vitro Scratch Assay
Scratch assay was performed in 6-well plates. To analyze cell migration, cells were transfected with miRNAs (or anti-miRNAs) or treated with chimera or aptamer alone for 72 hr. 72 hr post-transfection, the cells were seeded at a concentration of 5 × 105 cells. Treated cells were incubated again with chimera or aptamer and then treated with erlotinib. A scratched wound was scraped with a 20-μL tip in each well and then cells were continuously cultured in the media with 1% FBS for 144 hr. Microscopy images of the cultures were taken at 0, 24, 48, 72, and 144 hr, using a 5× objective.
Statistical Analysis
All experiments were performed at least three times. Continuous variables are given as mean ± SD. For comparisons between two groups, the Student’s t test was used to determine differences between mean values for normal distribution. Comparisons among more than three groups were determined by one-way ANOVA followed by Bonferroni’s post-hoc testing.
NSCLC patient survival was illustrated by Kaplan-Meier curves; survival differences between groups were examined with a log rank test. Analyses were conducted with GraphPad Prism 6 software. p values < 0.05 were considered statistically significant.
Author Contributions
V.R. and A.P. equally contributed to designing and performing all of the experiments and analyzing and interpreting the data; A.A. and S.N. performed the review experiments; D.F. gave technical support; F.P. and A.A. performed and analyzed all of the other experiments and gave technical support; M.G. and S.V. gave bioinformatics support; A.F. provided the biopsies of patients; C.L.E., G.R., and G.I contributed to the interpretation of data, providing advice; V.d.F. and G.C. conceived of and designed the study and analyzed and interpreted the data; G.C. provided financial support and wrote the paper; all the authors reviewed the manuscript.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This work was supported by funds from the Fondazione Berlucchi (G.C.), AIRC (14046 and 18473 to G.C.; 13345 and 9980 to V.d.F.), and CNR Flagship Project NanoMax (DESIRED) (to V.d.F.). We are grateful to Prof. Roberto Pacelli (University of Naples, Federico II) for his help in the radiosensitivity experiments.
Footnotes
Supplemental Information includes five figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.09.016.
Supplemental Information
References
- 1.Spira A., Ettinger D.S. Multidisciplinary management of lung cancer. N. Engl. J. Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 2.Yang P. Epidemiology of lung cancer prognosis: quantity and quality of life. Methods Mol. Biol. 2009;471:469–486. doi: 10.1007/978-1-59745-416-2_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sequist L.V., Martins R.G., Spigel D., Grunberg S.M., Spira A., Jänne P.A., Joshi V.A., McCollum D., Evans T.L., Muzikansky A. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J. Clin. Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z., Lee J.C., Lin L., Olivas V., Au V., LaFramboise T., Abdel-Rahman M., Wang X., Levine A.D., Rho J.K. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 8.Gaur A., Jewell D.A., Liang Y., Ridzon D., Moore J.H., Chen C., Ambros V.R., Israel M.A. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 9.Jiang J., Lee E.J., Gusev Y., Schmittgen T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Garofalo M., Condorelli G., Croce C.M. MicroRNAs in diseases and drug response. Curr. Opin. Pharmacol. 2008;8:661–667. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Navarro F., Lieberman J. miR-34 and p53: New Insights into a Complex Functional Relationship. PLoS ONE. 2015;10:e0132767. doi: 10.1371/journal.pone.0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y.L., Xu Y.J., Qiao C.W. MiR-34c-3p suppresses the proliferation and invasion of non-small cell lung cancer (NSCLC) by inhibiting PAC1/MAPK pathway. Int. J. Clin. Exp. Pathol. 2015;8:6312–6322. [PMC free article] [PubMed] [Google Scholar]
- 14.Hagman Z., Larne O., Edsjö A., Bjartell A., Ehrnström R.A., Ulmert D., Lilja H., Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int. J. Cancer. 2010;127:2768–2776. doi: 10.1002/ijc.25269. [DOI] [PubMed] [Google Scholar]
- 15.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 16.Peng D., Wang H., Li L., Ma X., Chen Y., Zhou H., Luo Y., Xiao Y., Liu L. miR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia. 2018;32:1180–1188. doi: 10.1038/s41375-018-0015-2. [DOI] [PubMed] [Google Scholar]
- 17.Catuogno S., Cerchia L., Romano G., Pognonec P., Condorelli G., de Franciscis V. miR-34c may protect lung cancer cells from paclitaxel-induced apoptosis. Oncogene. 2013;32:341–351. doi: 10.1038/onc.2012.51. [DOI] [PubMed] [Google Scholar]
- 18.Van Roosbroeck K., Calin G.A. Cancer Hallmarks and MicroRNAs: The Therapeutic Connection. Adv. Cancer Res. 2017;135:119–149. doi: 10.1016/bs.acr.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Catuogno S., Esposito C.L., Condorelli G., de Franciscis V. Nucleic acids delivering nucleic acids. Adv. Drug Deliv. Rev. 2018 doi: 10.1016/j.addr.2018.04.006. Published online April 6, 2018. [DOI] [PubMed] [Google Scholar]
- 20.Hermann T., Patel D.J. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 21.Banerjee J., Nilsen-Hamilton M. Aptamers: multifunctional molecules for biomedical research. J. Mol. Med. (Berl.) 2013;91:1333–1342. doi: 10.1007/s00109-013-1085-2. [DOI] [PubMed] [Google Scholar]
- 22.Ma H., Liu J., Ali M., Mahmood A., Labanieh L., Lu M., Iqbal S.M., Zhang Q., Zhao W., Wan Y. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem. Soc. Rev. 2015;44:1240–1256. doi: 10.1039/c4cs00357h. [DOI] [PubMed] [Google Scholar]
- 23.Cerchia L., De Franciscis V. Noncoding RNAs in cancer medicine. J. Biomed. Biotechnol. 2006;2006:73104. doi: 10.1155/JBB/2006/73104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farokhzad O.C., Cheng J., Teply B.A., Sherifi I., Jon S., Kantoff P.W., Richie J.P., Langer R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagalkot V., Farokhzad O.C., Langer R., Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. Engl. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 26.Wu X., Ding B., Gao J., Wang H., Fan W., Wang X., Zhang W., Wang X., Ye L., Zhang M. Second-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapy. Int. J. Nanomedicine. 2011;6:1747–1756. doi: 10.2147/IJN.S23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C.H., Dellamaggiore K.R., Ouellette C.P., Sedano C.D., Lizadjohry M., Chernis G.A., Gonzales M., Baltasar F.E., Fan A.L., Myerowitz R., Neufeld E.F. Aptamer-based endocytosis of a lysosomal enzyme. Proc. Natl. Acad. Sci. USA. 2008;105:15908–15913. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hicke B.J., Stephens A.W., Gould T., Chang Y.F., Lynott C.K., Heil J., Borkowski S., Hilger C.S., Cook G., Warren S., Schmidt P.G. Tumor targeting by an aptamer. J. Nucl. Med. 2006;47:668–678. [PubMed] [Google Scholar]
- 29.Tong G.J., Hsiao S.C., Carrico Z.M., Francis M.B. Viral capsid DNA aptamer conjugates as multivalent cell-targeting vehicles. J. Am. Chem. Soc. 2009;131:11174–11178. doi: 10.1021/ja903857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu T.C., Twu K.Y., Ellington A.D., Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N., Zhou C., Zhao J., Chen Y. Reversal of paclitaxel resistance in epithelial ovarian carcinoma cells by a MUC1 aptamer-let-7i chimera. Cancer Invest. 2012;30:577–582. doi: 10.3109/07357907.2012.707265. [DOI] [PubMed] [Google Scholar]
- 32.Cerchia L., Esposito C.L., Camorani S., Rienzo A., Stasio L., Insabato L., Affuso A., de Franciscis V. Targeting Axl with an high-affinity inhibitory aptamer. Mol. Ther. 2012;20:2291–2303. doi: 10.1038/mt.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catuogno S., Rienzo A., Di Vito A., Esposito C.L., de Franciscis V. Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J. Control. Release. 2015;210:147–159. doi: 10.1016/j.jconrel.2015.05.276. [DOI] [PubMed] [Google Scholar]
- 34.Esposito C.L., Cerchia L., Catuogno S., De Vita G., Dassie J.P., Santamaria G., Swiderski P., Condorelli G., Giangrande P.H., de Franciscis V. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol. Ther. 2014;22:1151–1163. doi: 10.1038/mt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iaboni M., Russo V., Fontanella R., Roscigno G., Fiore D., Donnarumma E., Esposito C.L., Quintavalle C., Giangrande P.H., de Franciscis V., Condorelli G. Aptamer-miRNA-212 Conjugate Sensitizes NSCLC Cells to TRAIL. Mol. Ther. Nucleic Acids. 2016;5:e289. doi: 10.1038/mtna.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackiewicz M., Huppi K., Pitt J.J., Dorsey T.H., Ambs S., Caplen N.J. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res. Treat. 2011;130:663–679. doi: 10.1007/s10549-011-1690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amarzguioui M., Rossi J.J. Principles of Dicer substrate (D-siRNA) design and function. Methods Mol. Biol. 2008;442:3–10. doi: 10.1007/978-1-59745-191-8_1. [DOI] [PubMed] [Google Scholar]
- 38.Okimoto R.A., Bivona T.G. AXL receptor tyrosine kinase as a therapeutic target in NSCLC. Lung Cancer (Auckl.) 2015;6:27–34. doi: 10.2147/LCTT.S60438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez S., Risolino M., Mandia N., Talotta F., Soini Y., Incoronato M., Condorelli G., Banfi S., Verde P. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34:3240–3250. doi: 10.1038/onc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garofalo M., Di Leva G., Romano G., Nuovo G., Suh S.S., Ngankeu A., Taccioli C., Pichiorri F., Alder H., Secchiero P. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Garofalo M., Romano G., Di Leva G., Nuovo G., Jeon Y.J., Ngankeu A., Sun J., Lovat F., Alder H., Condorelli G. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat. Med. 2011;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Romano G., Acunzo M., Garofalo M., Di Leva G., Cascione L., Zanca C., Bolon B., Condorelli G., Croce C.M. MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc. Natl. Acad. Sci. USA. 2012;109:16570–16575. doi: 10.1073/pnas.1207917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garofalo M., Quintavalle C., Di Leva G., Zanca C., Romano G., Taccioli C., Liu C.G., Croce C.M., Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27:3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 44.Acunzo M., Visone R., Romano G., Veronese A., Lovat F., Palmieri D., Bottoni A., Garofalo M., Gasparini P., Condorelli G. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai K.M., Bao X.L., Kong X.H., Jinag W., Mao M.R., Chu J.S., Huang Y.J., Zhao X.J. Hsa-miR-34c suppresses growth and invasion of human laryngeal carcinoma cells via targeting c-Met. Int. J. Mol. Med. 2010;25:565–571. doi: 10.3892/ijmm_00000378. [DOI] [PubMed] [Google Scholar]
- 46.Corney D.C., Hwang C.I., Matoso A., Vogt M., Flesken-Nikitin A., Godwin A.K., Kamat A.A., Sood A.K., Ellenson L.H., Hermeking H., Nikitin A.Y. Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallardo E., Navarro A., Viñolas N., Marrades R.M., Diaz T., Gel B., Quera A., Bandres E., Garcia-Foncillas J., Ramirez J., Monzo M. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 48.Thiel K.W., Giangrande P.H. Intracellular delivery of RNA-based therapeutics using aptamers. Ther. Deliv. 2010;1:849–861. doi: 10.4155/tde.10.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu F., Wang X., Li J., Gu K., Lv L., Zhang S., Che D., Cao J., Jin S., Yu Y. miR-34c-3p functions as a tumour suppressor by inhibiting eIF4E expression in non-small cell lung cancer. Cell Prolif. 2015;48:582–592. doi: 10.1111/cpr.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.