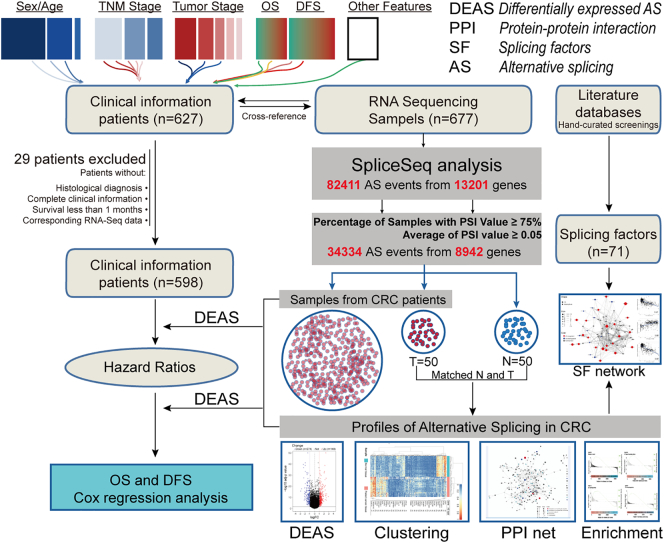
Fig. 1.
Flowchart for profiling the alternative splicing of CRC in a large-scale RNA-Seq data. RNA-Seq data and corresponding clinical information of CRC cohort were downloaded from the TCGA data portal. After excluding patients with incomplete clinical data and duplications, we combined these datasets into a large-scale CRC cohort, which was further analyzed by SpliceSeq. Through this process, PSI value was calculated for each AS events. Then, a series of stringent filters (Percentage of Samples with PSI Value ≥75, Average of PSI value ≥0.05) was implemented. Based on the PSI value of each AS events, we identified DEAS between CRC and normal tissues and further investigated the parent genes of these AS events by enrichment analysis. Next, the interaction network of these parent genes and regulation network of DEAS and splicing factors was visualized and analyzed. Finally, to assess the prognostic value of each DEAS event in CRC, their effect on OS and DFS was determined by Cox regression and Kaplan-Meier analysis.