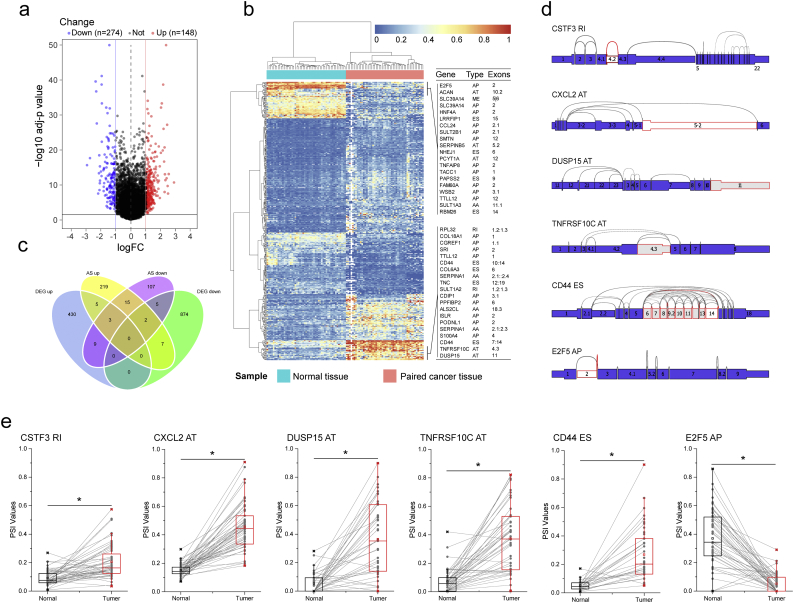
Fig. 3.
Identification of CRC-related aberrant AS. (a) The difference of AS events between paired colorectal cancer and paracancerous tissue. Volcano Plot visualizing the DEAS identified in CRC. The red and blue points in the plot represent the differentially expressed alternative splicing with statistical significance (adj P value <.05, |logFC| ≥ 1). (b) Heat map of the DEAS. The horizontal axis shows the clustering information of samples which were divided into two major clusters, and the clusters were adjacent normal tissue(N = 50) and paired tumor tissue(N = 50), respectively; the left longitudinal axis showed the clustering information of DEAS. The gradual change of color from green to red represents the expression of DEAS altered from low to high. (c) Venn diagram demonstrated the intersection set of DEAS and DEG. (d) The splice graph of some representative DEAS. The thin exon sections represent untranslated regions (UTR) and the thick exon sections represent coding regions. Exons are drawn to scale and the connecting arcs represent splice paths. (e) The difference of PSI value of AS events between primary CRC and corresponding adjacent normal tissues. Exon Skip (ES), Mutually Exclusive Exons (ME), Retained Intron (RI), Alternate Promoter (AP), Alternate Terminator (AT), Alternate Donor site (AD), and Alternate Acceptor site (AA). For Fig. 3a, t-test and BH correction was used for data analysis. For Fig. 3e, Student's t-test was used for data analysis. *: P < .0001 compared with adjacent normal tissue.