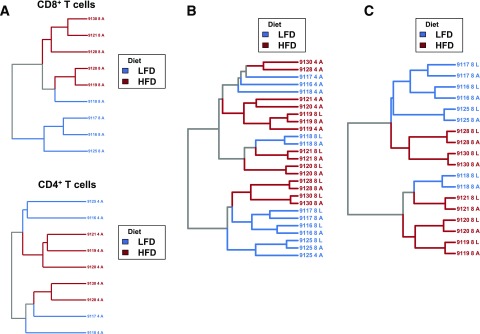
Figure 3.
Dietary group is predictive of repertoire similarity and synchronously alters the TCR repertoire in AT. Geometric mean overlap was used to calculate pairwise distance matrices on the basis of shared clonotypes with identical CDR3 amino acid sequences. Labels on each dendrogram correspond to a specific mouse, tissue type, and cell type (4 or 8). A: AT CD8+ (upper) and CD4+ repertoires (lower) share more TCRs within diet groups than between diet groups (P < 0.0001 for CD8+ and CD4+, permutation of pairwise distance matrix). This suggests that sets of shared TCRs are enriched and depleted in a diet-dependent fashion. B: Diet significantly affects repertoire similarity across multiple cell types and within multiple tissues (P = 0.01, permutation of pairwise distance matrix). TCRs shared between mice within multiple tissue and T-cell types distinguish between lean and obese mice. Diet-dependent repertoire similarity is observed in liver in addition to AT. C: CD8+ T-cell repertoires from liver and AT reflect simultaneous clonal expansion and sharing within each mouse and show that diet significantly affects repertoire similarity within the CD8+ compartment (P < 0.05, permutation of pairwise distance matrix). A, AT; L, liver.