Fig. 1.
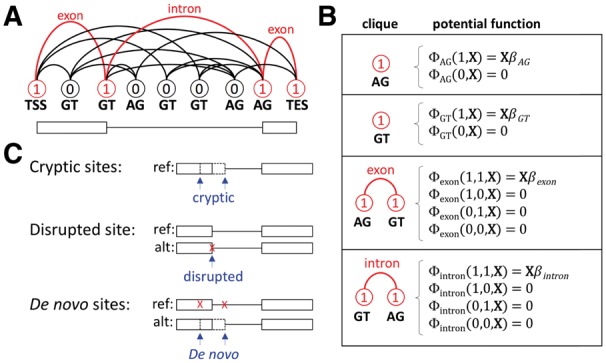
(A) A SGRF. Vertices denote splice sites, and edges denote exons and introns. A path from TSS to TES outlines a single gene structure. Labels 0 and 1 denote omission or inclusion, respectively, of a vertex on the selected path. (B) Cliques and their potential functions. SGRFs have only singleton and pair cliques. Potential functions for cliques labeled with any 0 do not contribute to the score, since they do not participate in the selected path. (C) Cryptic splice sites are unannotated splice sites near an annotated splice site. Disrupted splice sites exist in the reference but not in the alternate sequence. De novo splice sites exist in the alternate sequence but not in the reference
