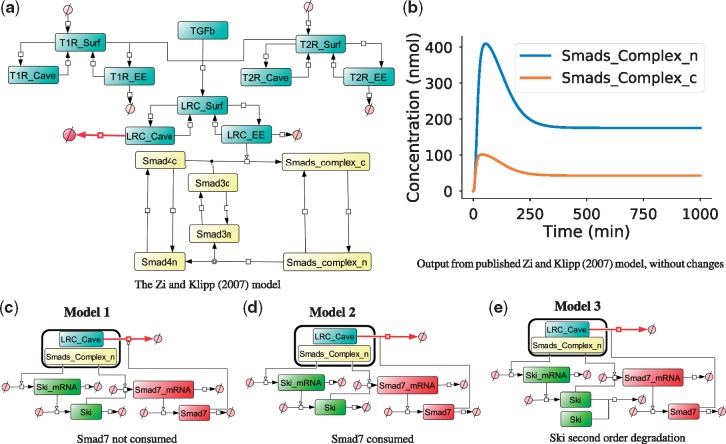
Fig. 1.
Network representation of ODE networks used in model selection problem. (a) The Zi and Klipp (2007) model is a common component of each model variant. (b) Simulation output from the Zi and Klipp (2007). (c–e) The model variable ‘Smads_Complex_n’ is responsible for transcription reactions in model variants while ‘LRC_Cave’ is degraded by Smad7 protein, thus completing the explicit representation of the Smad7 negative feedback loop. In (c) Model 1, Smad7 participates in but is not consumed by the reaction with LRC_Cave while in (d) Model 2, Smad7 is consumed by this process. In (e) Model 3, the same topology as Model 2 is assumed but it also incorporates second order mass action degradation kinetics for Ski protein