Abstract
Motivation
A reconciliation is an annotation of the nodes of a gene tree with evolutionary events—for example, speciation, gene duplication, transfer, loss, etc.—along with a mapping onto a species tree. Many algorithms and software produce or use reconciliations but often using different reconciliation formats, regarding the type of events considered or whether the species tree is dated or not. This complicates the comparison and communication between different programs.
Results
Here, we gather a consortium of software developers in gene tree species tree reconciliation to propose and endorse a format that aims to promote an integrative—albeit flexible—specification of phylogenetic reconciliations. This format, named recPhyloXML, is accompanied by several tools such as a reconciled tree visualizer and conversion utilities.
Availability and implementation
1 Introduction
The relationships between the history of genomes or species and the history of their constituent genes are often described through reconciliation. A reconciliation consists of an association between the nodes of a gene tree and the nodes or branches of a species tree, along with different evolutionary events undergone by the gene. For comprehensive reviews on the subject of reconciliations and their inference, see for example Nakhleh (2013) or Szöllősi et al. (2015).
Reconciliations can be used to understand the history of a specific gene family, and to study the evolutionary and functional relationships between several families. They can also be used to infer genome-wide parameters such as rates of gene duplication, loss, or lateral gene transfers (Sjöstrand et al., 2014; Szöllősi et al., 2013a), or population parameters such as divergence time and ancestral population size (Dutheil et al., 2009). Furthermore, reconciliation based metrics can be used as a criterion to construct better gene trees (Durand et al., 2006; Scornavacca et al., 2013; Sjöstrand et al., 2014; Szöllősi et al., 2013a; Wu et al., 2013) or better species tree (Boussau et al., 2013; Nakhleh, 2013).
There are many algorithms and software to infer reconciliations (Nakhleh, 2013; Szöllősi et al., 2015), and while they share many features, each has some unique characteristics.
Some methods work according to a parsimony principle [see for instance Durand et al. (2006), Bansal et al. (2012) and Chan et al. (2017)] while others rely on a probabilistic approach (Åkerborg et al., 2009; Sjöstrand et al., 2014; Szöllősi et al., 2013a). Reconciliation methods may differ in the type of events they consider. Some methods also require a dated species tree (i.e. a species tree where the relative timing of internal speciations is known) while others do not.
The fact that reconciliation programs (or rather each program family) use different formats to represent reconciliations makes it difficult to compare, switch between or use together reconciliations inferred from different pieces of software, which can hamper proper comparison and validation studies. This also means that any post-analysis or visualization software will either have limited scope (it will only be able to take as input the reconciliations of specific pieces of software) or be burdened by the implementation of readers for several formats.
In this paper, we aim to propose a generic reconciliation format encompassing the specificities of different reconciliation programs. This will make reconciliation based analysis more accessible to scientists without the need to develop or use multiple format conversion scripts.
In order to include all properties described in the scientific literature about gene tree species tree reconciliation, we should first be able to annotate gene tree nodes with events related to species tree nodes, such as speciations, and events associated to species tree branches, such as gene duplication (D), gene loss (L), lateral gene transfer (T), transfer with replacement (TR), gene conversion (C) and incomplete lineage sorting (ILS) (Mallo et al., 2014; Rasmussen and Kellis, 2012; Than et al., 2008). Reconciliations can be carried out with dated or undated species trees. In a dated species tree, the relative order of speciations is known and it would be desirable to be able to include information about the relative time at which the different events occurred in the reconciliation.
Transfers are written with two separate events: a gene lineage leaving a species tree branch (branching out) and then entering another species tree branch (transfer reception). As noted in Szöllősi et al. (2013b), most transfers originate from extinct or unsampled lineages (i.e. branches absent from the species tree). This implies that the bifurcation in the gene tree when a lineage leaves the species tree is not the transfer itself but actually a speciation toward an unsampled/extinct lineage. Our format nevertheless reflects the generality of this event by adopting a neutral label compatible with the different representations of transfers.
Moreover, this notion of evolution in unsampled lineages implies the possibility of a bifurcation in the gene tree in such a lineage. The children of the bifurcation can undergo transfers back to the sampled lineages. The unseen bifurcation might be a duplication, a speciation or a transfer between two unsampled lineage. Existing models are yet unable to discriminate these events. This idea is reflected in our format thanks to a specific way to specify a bifurcation in an unsampled lineage.
There have been previous attempts to develop formats able to represent evolutionary events along a phylogeny. The PhyloXML format (Han and Zmasek, 2009) is able to depict various annotations along a tree. It already has some way of representing evolutionary events along a phylogeny, but with limitations. For example PhyloXML lacks a mean to specify the species associated with the different events and only includes a rudimentary representation of transfers.
Adapting the already existing tags for evolutionary event in PhyloXML would require a near complete overhaul; rather, we propose a new format (recPhyloXML) with entirely new tags, ensuring no confusion with PhyloXML.
1.1 State of the art
Existing reconciliation formats can be broadly categorized in two groups.
The first group describes reconciliation events as labels in a Newick or NHX tree, in place of the nodal support (e.g. bootstrap) information or in a devoted NHX comment field. Programs like ALE (Szöllősi et al., 2013a), NOTUNG (Durand et al., 2006; Stolzer et al., 2012), DrML (Górecki and Eulenstein, 2014), phylotoo2 (Zheng and Zhang, 2017) or PrIME (Åkerborg et al., 2009; Sjöstrand et al., 2014) adhere to this group. The Newick-based reconciliation formats have the advantage of representing the phylogeny. However the reconciliation information often takes the space of other measures like bootstrap values [as in Szöllősi et al. (2013a), or Górecki and Eulenstein (2014)]. The NHX-based format solves this by allocating a specific space for the reconciliation. A common problem with NHX and Newick-based formats is that some characters are forbidden in the leaf names and annotations (These forbidden characters are : , : () ; [] in Newick and NHX.), while sometimes species or gene annotations contain these characters (whereas they rarely contain whole XML tags). In addition, there is no formal format for information contained in NHX comment fields; thus, this information may not be accessible across software platforms.
The second group represents reconciliations as lists of gene tree nodes mapping to species tree nodes, making references to an implicit or external gene tree (meaning that the gene tree structure might not be included in the reconciliation). Examples of such output formats are used by ranger-DTL (Bansal et al., 2012), ecceTERA (Jacox et al., 2016), DLcoalRecon (Rasmussen and Kellis, 2012), Mowgli (Doyon et al., 2010), the visualization software SylvX (Chevenet et al., 2016) or the simulation software SimPhy (Mallo et al., 2016).
2 Format presentation
To describe reconciliations, we present recPhyloXML, recGeneTreeXML, recSpeciesTreeXML, three grammars extending the PhyloXML format. We also introduce recGeoXML, a grammar to annotate reconciliations with geographic information.
They both rely on an XML structure composed of hierarchical tags. A specific tag may have different attributes which can be obligatory or optional.
In this section we briefly detail the structure of the PhyloXML used in our format. We then expand on the tags that are specific to reconciliations.
2.1 Elements in common with PhyloXML
In PhyloXML, a tree is delimited by the tag <phylogeny> </phylogeny>, which is included in a <phyloxml> </phyloxml> root tag that specifies that the file follows the PhyloXML format. Inside the <phylogeny> </phylogeny> tag, each clade is recursively inscribed in a <clade> </clade> tag. This clade tag possesses a facultative attribute to describe branch length. The name or identifier of the node is given in the <name> </name> tag. Further information can be included such as support value (<confidence></confidence>) or miscellaneous information (<description></description>).
2.2 New elements
In our format, a reconciliation (<recPhylo> tag) is defined as a set comprised of one or more reconciled gene trees (<recGeneTree> tag), and a species tree (<spTree> tag). These tags are described in the next section. Also, reconciled gene trees are always rooted and this is specified by using the tag
<phylogeny rooted=“true”></phylogeny>.
A recPhyloXML file allows you to store and share one or more reconciled genes trees and the associated species tree. A recGeneTreeXML file allows you to add a list of evolutionary events to the description of gene tree nodes (otherwise referred to as clades in PhyloXML), possibly also containing detailed geographic information thanks to the recGeoXML grammar (<geography> tag). This tag can also be used in a recSpeciesTreeXML file that currently differs from PhyloXML file only in this point.
2.3 recGeneTreeXML
recGeneTreeXML enriches the PhyloXML vocabulary by adding the complex tag <eventsRec> that must be included inside a <clade> tag.
The <eventsRec> tag contains the sequence of evolutionary events that occur along a gene tree branch.
Each type of evolutionary event is represented by a specific tag. These can be of two types, according to whether they concern a branch or a node of the gene tree:
Non terminal event:<transferBack>. This tag can be used as many times as necessary. This event does not cause any bifurcation in the gene tree.
Terminal events:<speciation>, <branchingOut>, <bifurcationOut>, <duplication>, <loss> and <leaf>. There is exactly one of these tags at the end of the sequence of events contained in the <eventsRec> tag.
Terminal events cause either a bifurcation in the gene tree (<speciation>, <branchingOut>, <bifurcationOut>, <duplication>) or the end of a lineage (<leaf>, <loss>).
Aside from the <bifurcationOut> and <transferBack> tags, all tags have an obligatory speciesLocation attribute that specifies in which species the event takes place. For <bifurcationOut>, the event always takes place in an unsampled/extinct lineage. <transferBack> events have instead a destinationSpecies attribute that specifies the species that receives the transfer. All event tags also have a facultative confidence attribute that is intended to store a support value for this event (Nguyen et al., 2013). Additionally, all event tags have a facultative timeSlice attribute that can, in models where the species tree is dated and subdivided for instance [as done for example in Doyon et al. (2010)], provide information on the timing of the event. Finally, the <leaf> tag has a facultative geneName attribute that can specify to which extant gene it corresponds. We now describe each event tag in details.
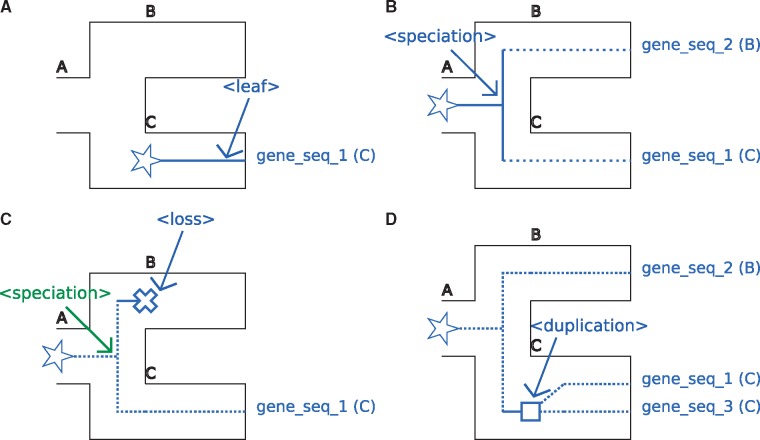
<leaf> tag:
The <leaf> tag indicates that the branch ends on a gene tree leaf; see Figure 1A. Note that the <leaf> tag also has a facultative geneName attribute that can specify to which extant gene it corresponds.
Fig. 1.
A. Representation of the <leaf> tag. B. Representation of the <speciation> tag. C. Representation of the <loss> tag. D. Representation of the <duplication> tag. The species tree is figured using black tube-like branches. The part of the gene tree the event occurs in is represented using plain lines. Additional parts of the gene tree are represented as dotted lines. Stars, squares and crosses respectively represent the beginning of a gene lineage, a gene duplication and a gene loss
Associated recGeneTreeXML code:
<clade>
<name>gene_seq_1</name>
<eventsRec>
<leaf speciesLocation=“C”></leaf>
</eventsRec>
</clade>
<speciation> tag:
The <speciation> tag describes a gene lineage undergoing a bifurcation due to a speciation; see Figure 1B.
Associated recGeneTreeXML code:
<clade>
<name>n1</name>
<eventsRec>
<speciation speciesLocation=“A”></speciation>
</eventsRec>
</clade>
<loss> tag:
The <loss> tag describes the loss of a gene copy and is a terminal tag (as with the <leaf> tag, there can be no tag following this one). Typically, it can follow a speciation event. See Figure 1C for an example.
Associated recGeneTreeXML code:
<!–Example with end tag <leaf>–>
<clade>
<name>n1</name>
<eventsRec>
<speciation>speciesLocation=“A”> </speciation>
</eventsRec>
<clade>
<name>gene_seq_1</name>
<eventsRec>
<leaf speciesLocation=“C”></leaf>
</eventsRec>
</clade>
<clade>
<name>LOST</name>
<eventsRec>
<loss speciesLocation=“B”></loss>
</eventsRec>
</clade>
</clade>
<duplication> tag:
The <duplication> tag represents a gene duplication inside a species tree branch; see Figure 1D.
Associated recGeneTreeXML code:
<clade>
<name>n1</name>
<eventsRec>
<duplication speciesLocation=“C”>
</duplication>
</eventsRec>
</clade>
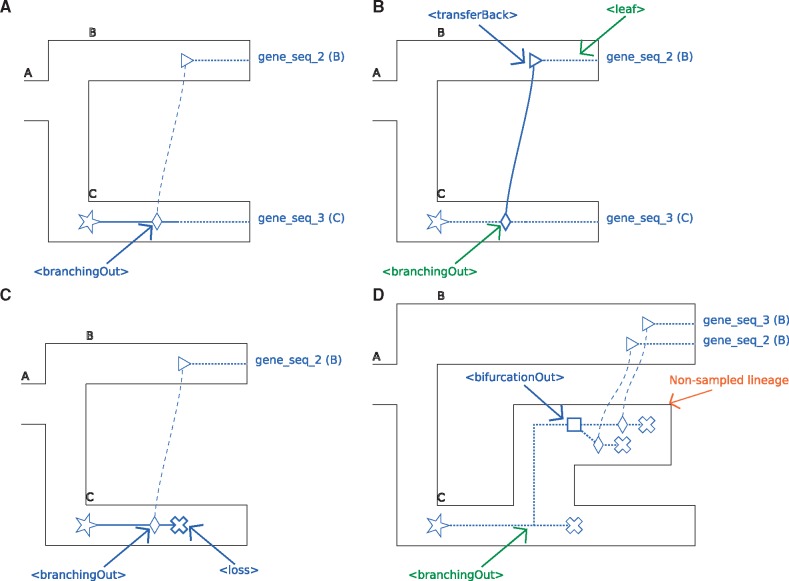
<branchingOut> tag:
The <branchingOut> tag represents an event where a gene lineage splits and one gene copy exits the species tree branch while the other gene copy remains in the species branch. It actually is the first step of an horizontal gene transfer event: a gene lineage leaving a species tree branch; see Figure 2A. Figure 2C also represents the case of a <branchingOut> where the child that remained in the same species was lost (<loss> tag).
Fig. 2.
A. Representation of the <branchingOut> tag. B. Representation of the <transferBack> tag. C. Representation of a <branchingOut> tag followed by a <loss> tag. D. Representation of the <bifurcationOut> tag. These figure uses the same conventions as Figure 1 with the following additions. For the <bifurcationOut> tag (D.), which is specific of the model of Szöllősi et al. (2013b), extinct/unsampled lineages are represented as branches of the species tree that do not extend all the way to the right. Diamonds and triangles respectively represent a transfer leaving and entering a branch of the species tree (note that the transfers leave a branch of the species tree that corresponds to an extinct/unsampled lineage)
Associated recGeneTreeXML code:
<clade>
<name>n1</name>
<eventsRec>
<branchingOut speciesLocation=“C”>
</branchingOut>
</eventsRec>
</clade>
<transferBack> tag:
The <transferBack> tag represents an horizontal gene transfer toward a branch of the species tree; see Figure 2B.
Associated recGeneTreeXML code:
<!–Example with end tag <leaf> –>
<clade>
<name>gene_seq_2</name>
<eventsRec>
<transferBack destinationSpecies=“B”>
</transferBack>
<leaf speciesLocation=“B”></leaf>
</eventsRec>
</clade>
<bifurcationOut> tag:
The <bifurcationOut> tag represents a bifurcation in the species tree that would happen while the gene evolves along an unsampled/extinct species (i.e. one that is not represented in the species tree, see the <branchingOut> and <transferBack> tags above); see Figure 2D.
Associated recGeneTreeXML code:
<clade>
<name>n1</name>
<eventsRec>
<bifurcationOut></bifurcationOut>
</eventsRec>
</clade>
2.4 Note on the lateral gene transfer representation
A lateral gene transfer is represented in two steps: one that specifies the species where the transfer originates, and the other that specifies the species receiving the transfer. These two successive steps are respectively represented by the <branchingOut> and <transferBack> tags. See the different parts of Figure 2, along with Figures 3 and 4 for illustrations of these concepts.
Fig. 3.
A <recPhylo> object containing a species tree and one reconciled gene tree
Fig. 4.
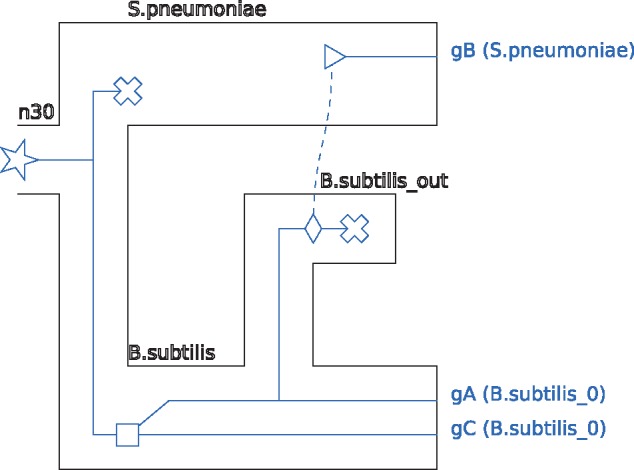
A visualization of the reconciled gene tree of Figure 3
2.5 recGeoXML
Geographical annotations can be indicated for gene and species tree nodes thanks to the <geography> tag. Such an annotation mainly consists in an area, KML information for displaying areas in GIS software and geographic information as defined in the usual PhyloXML grammar. An area (<area>) is specified by a name, a description, a value such as a support and a source (e.g. ‘observed’ or ‘inferred by Beast’).
2.6 recPhyloXML
recPhyloXML facilitates the packaging of several gene families reconciled to the same species tree. Its structure is fairly simple. A <recPhylo> root tag contains the following sequence:
0 to 1 species tree in recSpeciesTreeXML format, contained in the <spTree> tag.
1 to n gene family trees in recGeneTreeXML format, each defined in a separate <recGeneTree> tag.
<!– skeleton of a recphylo object with a species tree and two reconciled gene trees –>
<recPhylo>
<spTree>
<!– recSpeciesTreeXML species tree –>
…
</spTree>
<recGeneTree>
<!– first reconciled gene tree –>
…
</recGeneTree>
<recGeneTree>
<!– second reconciled gene tree –>
…
</recGeneTree>
</recPhylo>
A complete example of a <recPhylo> object containing a species tree and a reconciled gene tree can be seen in Figure 3 and a visualization of this reconciled gene tree can be seen in Figure 4.
3 Availability
A detailed description of the recPhyloXML format, as well as a schema definition file (This is a file formally describing the format, used by many XML tools.), is available at http://phylariane.univ-lyon1.fr/recphyloxml/. This website also presents a tool that can generate a visual representation of any reconciled tree or group of reconciled trees in the recPhyloXML format. The generated file is a.svg file, which easily allows for further manipulation, like changing the color scheme. This tool has been used to generate the basis for the figures showing reconciled gene trees in this manuscript.
The recPhyloXML format has already been implemented as an output option in the reconciliation software ALE (Szöllősi et al., 2013a), in the Treerecs software [https://gitlab.inria.fr/Phylophile/Treerecs, this program corrects gene trees with a species tree using principles described in Noutahi et al. (2016) and Lafond et al. (2012)], in the reconciliation web server http://phylotoo2.appspot.com/ (Zheng and Zhang, 2017), in the genome simulation software Zombi (https://github.com/AADavin/ZOMBI), both as input and output options in the adjacency history computing software DeCoSTAR (Duchemin et al., 2017).
Furthermore, scripts have been developed to convert the reconciliations produced by ecceTERA (Jacox et al., 2016), NOTUNG (Durand et al., 2006) and PrIME (Åkerborg et al., 2009) into recPhyloXML, and a script for converting reconciliations produced by RANGER-DTL (Bansal et al., 2018) is currently under development. Additional scripts are also available to convert a recPhyloXML reconciled tree in the Newick format, count the different events represented in a recPhyloXML file, combine different files into one or extract specific trees from a file. APIs have been written to import and export in recPhyloXML for the C++ library Bio++ (Gueguen et al., 2013), for the python libraries ETE3 (Huerta-Cepas et al., 2016) and Biopython (Cock et al., 2009). All these scripts and APIs are available at https://github.com/WandrilleD/recPhyloXML.
4 Conclusion
With the growing number of available reconciliation models and pieces of software, it becomes crucial to be able to exchange and compare their results. recPhyloXML is a format that can accommodate many reconciliation features (dated/undated; with or without lateral gene transfers). It relies on an XML structure which is a standard format for nested data that already has multiple API libraries in various programming languages. We provide a detailed description of the recPhyloXML format on a website, along with a tool to visualize it.
We designed the format to be flexible in order to be able to create extensions that allow the representation of different forms of reconciliations. We are planning for future extensions for the format that would include a representation of the coalescent process that underlies ILS. recPhyloXML could also be extended to support gene conversion by a paralog or horizontal gene transfer with replacement.
Acknowledgements
The authors would like to thank Dannie Durand, Han Lai and Maureen Stolzer for their advice on both the format and the manuscript.
Funding
This work was funded by the Agence Nationale pour la Recherche, Ancestrome project ANR-10-BINF-01-01 and by the Institut de Biologie Computationnelle ANR-11-BINF-0002. G.J.Sz., W.D. and A.A.D. received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 714774. C.D and D.D. were funded by Swiss National Science Foundation grant 150654.
Conflict of Interest: none declared.
References
- Åkerborg Ö. et al. (2009) Simultaneous Bayesian gene tree reconstruction and reconciliation analysis. Proc. Natl. Acad. Sci. USA, 106, 5714–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M.S. et al. (2012) Efficient algorithms for the reconciliation problem with gene duplication, horizontal transfer and loss. Bioinformatics, 28, i283–i291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M.S. et al. (2018) Ranger-dtl 2.0: Rigorous reconstruction of gene-family evolution by duplication, transfer, and loss. Bioinformatics, 34, 3214–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussau B. et al. (2013) Genome-scale coestimation of species and gene trees. Genome Res., 23, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y-b. et al. (2017) Inferring incomplete lineage sorting, duplications, transfers and losses with reconciliations. J. Theor. Biol., 432, 1–13. [DOI] [PubMed] [Google Scholar]
- Chevenet F. et al. (2016) SylvX: a viewer for phylogenetic tree reconciliations. Bioinformatics, 32, 608–610. [DOI] [PubMed] [Google Scholar]
- Cock P.J.A. et al. (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics, 25, 1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon J.P. et al. (2010) An efficient algorithm for gene/species trees parsimonious reconciliation with losses, duplications, and transfers. In: Proceedings of the 2010 International Conference on Comparative Genomics, RECOMB-CG’10, pp. 93–108.
- Duchemin W. et al. (2017) Decostar: reconstructing the ancestral organization of genes or genomes using reconciled phylogenies. Genome Biol. Evol, 9, 1312–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand D. et al. (2006) A hybrid micro-macroevolutionary approach to gene tree reconstruction. J. Comput. Biol., 13, 320–335. [DOI] [PubMed] [Google Scholar]
- Dutheil J.Y. et al. (2009) Ancestral population genomics: the coalescent hidden Markov Model Approach. Genetics, 183, 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górecki P., Eulenstein O. (2014) DrML: probabilistic modeling of gene duplications. J. Comput. Biol., 21, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen L. et al. (2013) Bio++: efficient extensible libraries and tools for computational molecular evolution. Mol. Biol. Evol., 30, 1745–1750. [DOI] [PubMed] [Google Scholar]
- Han M.V., Zmasek C.M. (2009) phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics, 10, 356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J. et al. (2016) ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol., 33, 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacox E. et al. (2016) ecceTERA: comprehensive gene tree-species tree reconciliation using parsimony. Bioinformatics32, 2056–2058. [DOI] [PubMed] [Google Scholar]
- Lafond M. et al. (2012). An optimal reconciliation algorithm for gene trees with polytomies. In: WABI, pp. 106–122.
- Mallo D. et al. (2014) Unsorted homology within locus and species trees. Syst. Biol., 63, 988–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo D. et al. (2016) SimPhy: phylogenomic simulation of gene, locus and species trees. Syst. Biol., 65, 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhleh L. (2013) Computational approaches to species phylogeny inference and gene tree reconciliation. Trends Ecol. Evol., 28, 719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.H. et al. (2013) Support measures to estimate the reliability of evolutionary events predicted by reconciliation methods. PLoS One, 8, e73667–e73614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutahi E. et al. (2016) Efficient gene tree correction guided by genome evolution. PLoS One, 11, e0159559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M.D., Kellis M. (2012) Unified modeling of gene duplication, loss, and coalescence using a locus tree. Genome Res., 22, 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scornavacca C. et al. (2013) Representing a set of reconciliations in a compact way. J. Bioinf. Comput. Biol., 11, 1250025.. [DOI] [PubMed] [Google Scholar]
- Sjöstrand J. et al. (2014) A bayesian method for analyzing lateral gene transfer. Syst. Biol., 63, 409–420. [DOI] [PubMed] [Google Scholar]
- Stolzer M. et al. (2012) Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics, 28, i409–i415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllősi G.J. et al. (2013a) Efficient exploration of the space of reconciled gene trees. Syst. Biol., 62, 901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllősi G.J. et al. (2013b) Lateral gene transfer from the dead. Syst. Biol., 62, 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllősi G.J. et al. (2015) The inference of gene trees with species trees. Syst. Biol., 64, e42–e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than C. et al. (2008) Phylonet: a software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinformatics, 9, 322.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.-C. et al. (2013) TreeFix: statistically informed gene tree error correction using species trees. Syst. Biol., 62, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhang L. (2017) Reconciliation with non-binary gene trees revisited. J. ACM, 64, 1–28. [Google Scholar]