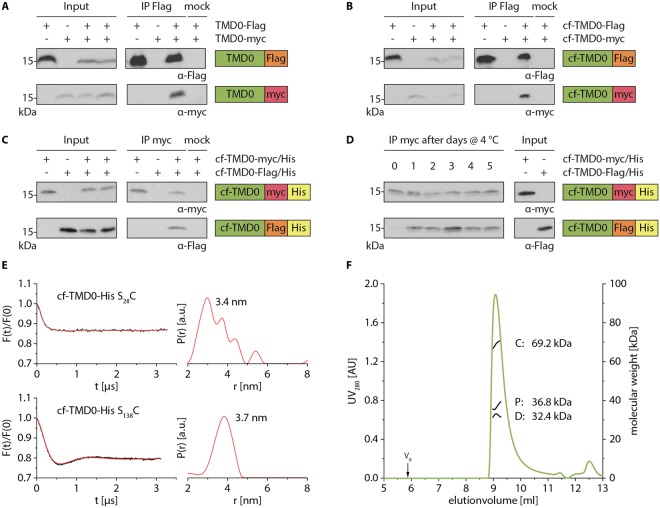
Figure 3.
TMD0 oligomerization. (A,B) To test oligomerization of TMD0 in cellular environment, HEK 293 T cells were transiently transfected with wt (A) and cf variants (B) of TMD0 carrying a C-terminal Flag- or myc-tag. After solubilization with 1% digitonin, co-immunoprecipitation with Flag-tag specific antibody or isotype control (mock) was performed and analyzed by immunoblotting. Input represents 1/25 aliquot of the volume used for immunoprecipitation. Original, uncropped immunoblots are shown in supplementary information. (C) To analyze oligomerization of cf expressed TMD0, precipitates of separately produced cf-TMD0-myc/His and cf-TMD0-Flag/His were mixed, solubilized in LMPG, purified and reconstituted in c6-DHPC. Single or co-purified TMD0 variants (input) were immunoprecipitated by myc-tag specific antibody or isotype control and analyzed by immunoblotting. Input represents 1/15 aliquot of the volume used for pull-down. Original, uncropped immunoblots are shown in supplementary information. (D) To probe the oligomerization dynamics, cf expressed TMD0 variants were separately purified and refolded (input). Cf-TMD0-myc/His and cf-TMD0-Flag/His were mixed in a 1:1 molar ratio and incubated at 4 °C up to five days. Exchange of subunits was analyzed by immunoprecipitation with myc-tag specific antibody. Input represents 1/15 aliquot of the volume used for pull-down. Original, uncropped immunoblots are shown in supplementary information. (E) Oligomerization of MTSL labeled cf-TMD0-His S28C and cf-TMD0-His S138C was investigated by PELDOR spectroscopy. PELDOR traces were background corrected (left), and corresponding distance distributions were calculated by Tikhonov regularization (right). (F) Dimer formation of cf-TMD0-His was determined by SEC-MALS. 400 µg of purified cf-TMD0-His were applied on a 15 ml TSK-Gel G3000 SWXL column (green line). Molecular weight distributions were calculated by three-detector method using Astra software and are depicted in black lines (C: protein-detergent conjugate, P: protein, D: detergent).