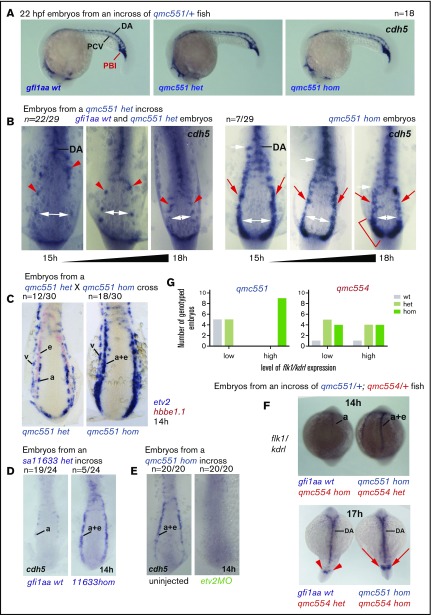
Figure 5.
Gfi1aa suppresses the endothelial gene expression program in primitive erythroblasts developing from the posterior lateral mesoderm. The figure shows zebrafish embryos stained in WISH experiments. Lateral views of embryos are shown in panel A. Flat-mounts of the posterior parts of embryos are displayed in panels B-E. Posterior and dorsal views of embryos are presented in the top and bottom panels of F, respectively. (A) Wt, gfi1aaqmc551het, and gfi1aaqmc551hom embryos display very similar cdh5 expression patterns at 22 hpf. Embryos were stained, photographed, and genotyped; 4 of 18 embryos were gfi1aaqmc551hom. Representative wt, gfi1aaqmc551het, and gfi1aaqmc551hom embryos are shown. (B) The expression pattern of the endothelial gene cdh5 is expanded in the posterior lateral mesoderm of Gfi1aa-depleted embryos. A mix of 15 to 18 hpf embryos derived from gfi1aaqmc551het incross were stained for the expression of cdh5, flat-mounted, photographed, and genotyped. Embryo images were sorted on the basis of the level of cdh5 expression and then arranged according to their development stage, which was judged by the distance between the bilateral PLM stripes. In younger embryos, this distance is larger than in older embryos (see white double arrows). Genotyping revealed that all gfi1aaqmc551hom mutant embryos displayed expanded expression patterns for cdh5 in the PLM when compared with wt and gfi1aaqmc551het embryos. The white arrows indicate the anterior extent of the solid stripe of cdh5+ cells in the PLM. Red arrowheads point at normal cdh5 gene expression in dispersed small clusters of PLM cells in wt and gfi1aaqmc551het embryos.28,81 Red arrows highlight unusually robust cdh5 expression in a solid stripe of PLM cells of gfi1aaqmc551hom mutant embryos. A red bracket indicates robust cdh5 expression in the posterior part of the PLM in the 18 hpf gfi1aaqmc551hom embryo. (C) In the absence of Gfi1aa, mRNA for the endothelial transcription factor Etv2 is ectopically expressed in prRBCs. Embryos from a double WISH experiment are shown. Embryos were first stained for the presence of etv2 mRNA in purple, and subsequently with a probe for hbbe1.1 mRNA in red. In the gfi1aaqmc551het embryos, etv2 is expressed in 2 stripes of EC progenitors, arterial (a) progenitors medially and venous (v) progenitors laterally. This is consistent with previous findings in wt embryos.25 Please note that the hbbe1.1-expressing primitive erythroid (e) progenitors are lateral to and much more closely associated with the arterial EC progenitors. In the gfi1aaqmc551hom mutant embryos, the medial stripe of etv2+ cells is wider and encompasses arterial EC progenitors and prRBCs (a+e). Please note that the purple staining in prRBCs masks any red staining that may be present. (D) The cdh5 expression pattern is expanded in gfi1aasa11633 homozygous mutant embryos. Embryos were stained and genotyped. Representative embryos are shown. (E) Endothelial and erythroid cdh5 expression require the presence of the endothelial TF Etv2. Two- and 4-cell stage embryos from a gfi1aaqmc551hom incross were injected with 10 ng of a previous validated etv2 5′ untranslated region (UTR) MO.63,64 Embryos were collected at 14 hpf and stained by WISH. None of the morphant embryos showed any staining. An 18-hpf wt embryo that had been added to the batch of morphant embryos before the WISH experiment showed normal cdh5 expression in ECs (data not shown), confirming that the WISH procedure successfully detected cdh5 mRNA. (F-G) The level of expression of the EC marker flk1/kdrl in the posterior lateral mesoderm correlates with the genotype of gfi1aa, and not with the genotype of gfi1b. In this experiment, gfi1aaqmc551/gfi1bqmc554 double-heterozygous fish were incrossed. Their embryos were stained by WISH, subsequently photographed, and genotyped. Representative embryos with weak and strong staining are shown. The bar charts in (G) summarize the collective results of the gfi1aa and gfi1b genotyping performed on 14- and 17-hpf somite stage embryos that displayed low or high levels of flk1/kdrl expression in the PLM. The flk1/kdrl data support the view that the EC gene upregulation is transient in gfi1aa single- and gfi1aa/1b double-mutant embryos.