Fig. 1.
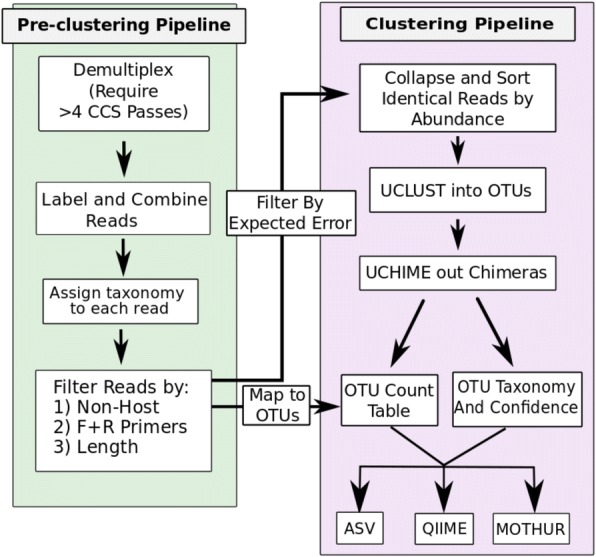
Overview of the MCSMRT pipeline represented as a flowchart. MCSMRT analysis of 16S rRNA reads from the PacBio is carried out in two steps: In the pre-clustering step, CCS reads are generated during demultiplexing, labeled by sample, pooled together, and then filtered based on several criteria (length distribution, terminal matches to the primer sequences, and not aligning to a provided host or background genome sequence). Before the clustering step, CCS reads are filtered based on cumulative expected error (EE < 1). The clustering pipeline uses UCLUST to identify and sort unique sequences based on their abundance, clusters CCS reads into OTUs (filtering out chimeric reads during clustering), and then using uchime after clustering as a second chimera removal step. An OTU count table is created by mapping the filtered results from the end of the pre-clustering pipeline, and each OTU is taxonomically classified based on a representative “centroid” sequence. Taxonomic classification is also applied to all filtered reads, and ASV detection by MED can be applied on multiple alignments of sets of related sequencing, grouped by either OTU or binned by taxonomic level
